1A2P
 
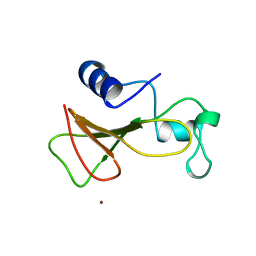 | | BARNASE WILDTYPE STRUCTURE AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | BARNASE, ZINC ION | | Authors: | Martin, C, Richard, V, Salem, M, Hartley, R.W, Mauguen, Y. | | Deposit date: | 1998-01-07 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refinement and structural analysis of barnase at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1KEN
 
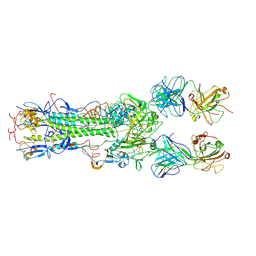 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH AN ANTIBODY THAT PREVENTS THE HEMAGGLUTININ LOW PH FUSOGENIC TRANSITION | | Descriptor: | alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin HA1, hemagglutinin HA2, ... | | Authors: | Barbey-Martin, C, Gigant, B, Bizebard, T, Calder, L.J, Wharto, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 2001-11-16 | | Release date: | 2002-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An Antibody that Prevents the Hemagglutinin Low pH Fusogenic Transition
VIROLOGY, 294, 2002
|
|
8QOT
 
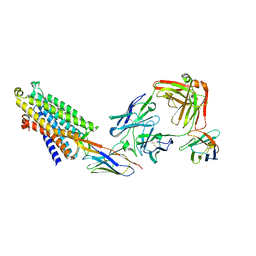 | | Structure of the mu opioid receptor bound to the antagonist nanobody NbE | | Descriptor: | Anti-Fab Nanobody, Mu-type opioid receptor, NabFab HC, ... | | Authors: | Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Manglik, A, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of mu-Opioid Receptor-Targeting by a Nanobody Antagonist.
Biorxiv, 2023
|
|
8WKY
 
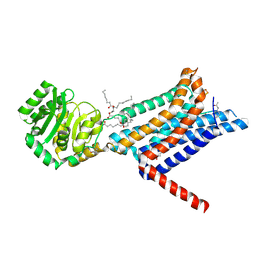 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S25 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, N-(2-aminoethyl)-5-(2-{[4-(morpholin-4-yl)pyridin-2-yl]amino}-1,3-thiazol-5-yl)pyridine-3-carboxamide, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
8WKZ
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with S31 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Gimenez, L.E, Martin, C, Yu, J, Hollanders, C, Hernandez, C, Dahir, N.S, Wu, Y, Yao, D, Han, G.W, Wu, L, Poorten, O.V, Lamouroux, A, Mannes, M, Tourwe, D, Zhao, S, Stevens, R.C, Cone, R.D, Ballet, S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Cocrystal Structures of Peptide Antagonists Bound to the Human Melanocortin Receptor 4 Unveil Unexplored Grounds for Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
6YBA
 
 | | HAdV-F41 Capsid | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Perez Illana, M, Martinez, M, Mangroo, C, Brown, M, Marabini, R, San Martin, C. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses.
Sci Adv, 7, 2021
|
|
9EMA
 
 | | RUVBL1/2 in complex with ATP and CB-6644 inhibitor | | Descriptor: | 5-chloranyl-2-ethoxy-4-fluoranyl-~{N}-[4-[[3-(methoxymethyl)-1-oxidanylidene-6,7-dihydro-5~{H}-pyrazolo[1,2-a][1,2]benzodiazepin-2-yl]amino]-2,2-dimethyl-4-oxidanylidene-butyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Perrote, A, Llorca, O, Garcia-Martin, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanism of allosteric inhibition of RUVBL1-RUVBL2 by the small-molecule CB-6644
Cell Rep Phys Sci, 2024
|
|
9EMC
 
 | |
1W8X
 
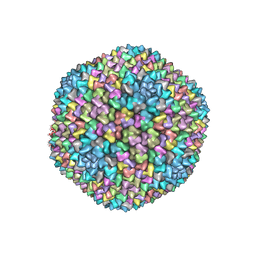 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1GW7
 
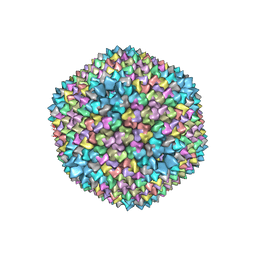 | | QUASI-ATOMIC RESOLUTION MODEL OF BACTERIOPHAGE PRD1 CAPSID, OBTAINED BY COMBINED CRYO-EM AND X-RAY CRYSTALLOGRAPHY. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | San Martin, C, Huiskonen, J, Bamford, J.K.H, Butcher, S.J, Fuller, S.D, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-03-08 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Minor Proteins, Mobile Arms, and Membrane-Capsid Interactions in Bacteriophage Prd1 Capsid Assembly
Nat.Struct.Biol., 9, 2002
|
|
8V8K
 
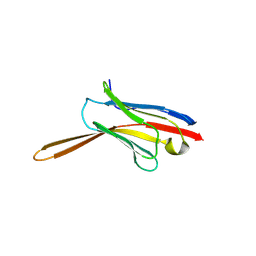 | | Crystal Structure of Nanobody NbE | | Descriptor: | Nanobody NbE | | Authors: | Koehl, A, Manglik, A, Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Steyaert, J, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-12-05 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of mu-opioid receptor targeting by a nanobody antagonist.
Nat Commun, 15, 2024
|
|
6PT2
 
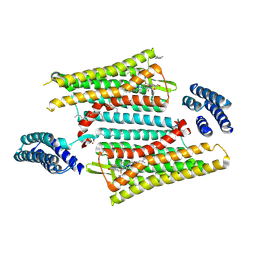 | | Crystal structure of the active delta opioid receptor in complex with the peptide agonist KGCHM07 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Delta opioid receptor, ... | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
1GW8
 
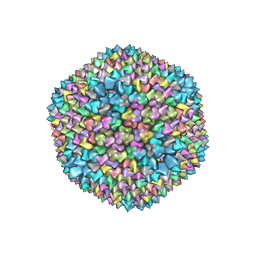 | | quasi-atomic resolution model of bacteriophage PRD1 sus607 mutant, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | San Martin, C, Huiskonen, J, Bamford, J.K.H, Butcher, S.J, Fuller, S.D, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2002-03-08 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.3 Å) | | Cite: | Minor Proteins, Mobile Arms and Membrane-Capsid Interactions in the Bacteriophage Prd1 Capsid.
Nat.Struct.Biol., 9, 2002
|
|
6PT3
 
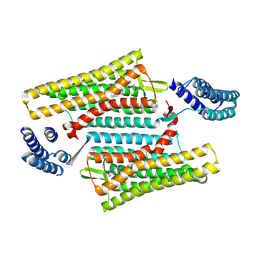 | | Crystal structure of the active delta opioid receptor in complex with the small molecule agonist DPI-287 | | Descriptor: | 4-[(R)-[(2S,5R)-4-benzyl-2,5-dimethylpiperazin-1-yl](3-hydroxyphenyl)methyl]-N,N-diethylbenzamide, Delta opioid receptor | | Authors: | Claff, T, Yu, J, Blais, V, Patel, N, Martin, C, Wu, L, Han, G.W, Holleran, B.J, Van der Poorten, O, Hanson, M.A, Sarret, P, Gendron, L, Cherezov, V, Katritch, V, Ballet, S, Liu, Z, Muller, C.E, Stevens, R.C. | | Deposit date: | 2019-07-14 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Elucidating the active delta-opioid receptor crystal structure with peptide and small-molecule agonists.
Sci Adv, 5, 2019
|
|
7B00
 
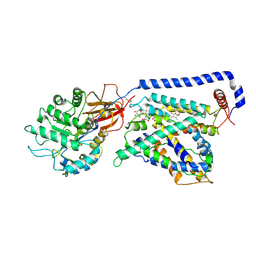 | | Human LAT2-4F2hc complex in the apo-state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Isoform 2 of 4F2 cell-surface antigen heavy chain, ... | | Authors: | Rodriguez, C.F, Escudero-Bravo, P, Garcia-Martin, C, Boskovic, J, Errasti-Murugarren, E, Palacin, M, Llorca, O. | | Deposit date: | 2020-11-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for substrate specificity of heteromeric transporters of neutral amino acids.
Proc Natl Acad Sci U S A, 118, 2021
|
|
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
5BMN
 
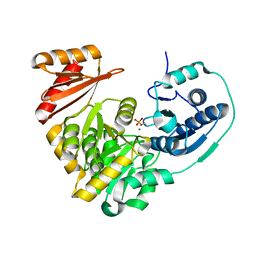 | | Crystal Structure of APO form of Phosphoglucomutase from Xanthomonas citri | | Descriptor: | MAGNESIUM ION, Phosphoglucomutase | | Authors: | Goto, L.S, Pereira, H.M, Novo Mansur, M.T.M, Brandao-Neto, J. | | Deposit date: | 2015-05-22 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural and functional characterization of the phosphoglucomutase from Xanthomonas citri subsp. citri.
Biochim.Biophys.Acta, 1864, 2016
|
|
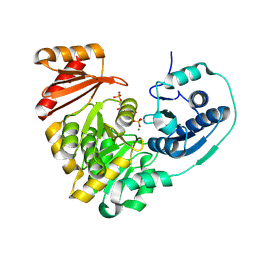
5BMP
 
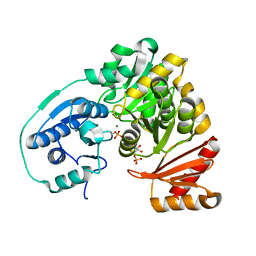 | |
5KL0
 
 | | Crystal Structure of Phosphoglucomutase from Xanthomonas citri citri complexed with Glucose-1,6-biphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Goto, L.S, Pereira, H.M, Novo Mansur, M.T.M. | | Deposit date: | 2016-06-23 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Structural and functional characterization of the phosphoglucomutase from Xanthomonas citri subsp. citri.
Biochim.Biophys.Acta, 1864, 2016
|
|
8BE6
 
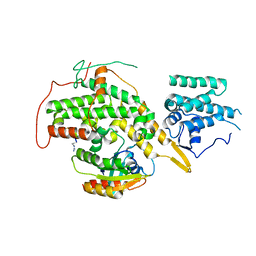 | | Crystal structure of SOS1-HRas-peptidomimetic2 | | Descriptor: | GTPase HRas, SOS1-HRas-peptidomimetic2, Son of sevenless homolog 1 | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.89880252 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
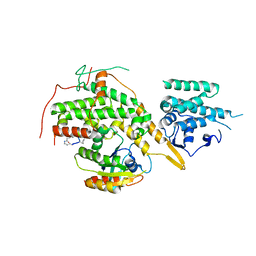
8BE7
 
 | |
8BE8
 
 | | Crystal structure of SOS1-HRas-peptidomimetic4 | | Descriptor: | FORMIC ACID, GTPase HRas, SOS1-HRas-peptidomimetic4, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BE9
 
 | | Crystal structure of SOS1-HRas-peptidomimetic5 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GTPase HRas, ... | | Authors: | Fischer, B, Wohlkonig, A, Steyaert, J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Nanobody Loop Mimetics Enhance Son of Sevenless 1-Catalyzed Nucleotide Exchange on RAS.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8BEA
 
 | |
6UPA
 
 | | Crystal Structure of GTPase Domain of Human Septin 2/Septin 6 Heterocomplex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rosa, H.V.D, Brandao-Neto, J, Martins, C, Araujo, A.P.U, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular Recognition at Septin Interfaces: The Switches Hold the Key.
J.Mol.Biol., 432, 2020
|
|
