2LSN
 
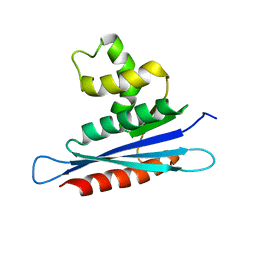 | | Solution structure of PFV RNase H domain | | Descriptor: | RNase H | | Authors: | Leo, B, Schweimer, K, Woehrl, B. | | Deposit date: | 2012-05-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the prototype foamy virus RNase H domain indicates an important role of the basic loop in substrate binding.
Retrovirology, 9, 2012
|
|
5MZN
 
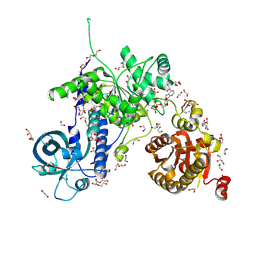 | | Helicase Sen1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Leonaite, B, Basquin, J, Conti, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-19 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family.
EMBO J., 36, 2017
|
|
6B0G
 
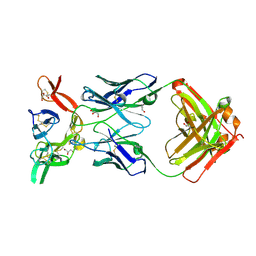 | | Crystal structure of Pfs25 in complex with the transmission blocking antibody 1245 | | Descriptor: | 1245 Antibody, Heavy Chain, Light Chain, ... | | Authors: | McLeod, B, Scally, S.W, Bosch, A, King, C.R, Julien, J.P. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular definition of multiple sites of antibody inhibition of malaria transmission-blocking vaccine antigen Pfs25.
Nat Commun, 8, 2017
|
|
6B0H
 
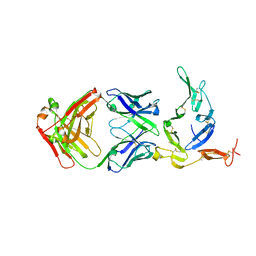 | | Crystal structure of Pfs25 in complex with the transmission blocking antibody 1262 | | Descriptor: | 1,2-ETHANEDIOL, 1262 antibody, heavy chain, ... | | Authors: | McLeod, B, Scally, S.W, Bosch, A, King, C.R, Julien, J.P. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular definition of multiple sites of antibody inhibition of malaria transmission-blocking vaccine antigen Pfs25.
Nat Commun, 8, 2017
|
|
7UNB
 
 | | Crystal structure of malaria transmission-blocking antigen Pfs48/45-6C variant in complex with human antibodies RUPA-117 and RUPA-47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Gametocyte surface protein P45/48, RUPA-117 Fab heavy chain, ... | | Authors: | Hailemariam, S, McLeod, B, Julien, J.-P. | | Deposit date: | 2022-04-10 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Vaccination with a structure-based stabilized version of malarial antigen Pfs48/45 elicits ultra-potent transmission-blocking antibody responses.
Immunity, 55, 2022
|
|
4TZ4
 
 | | Crystal Structure of Human Cereblon in Complex with DDB1 and Lenalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Lenalidomide, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B, Riley, M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
To Be Published
|
|
4TZC
 
 | | Crystal Structure of Murine Cereblon in Complex with Thalidomide | | Descriptor: | Protein cereblon, S-Thalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
3COK
 
 | | Crystal structure of PLK4 kinase | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, Serine/threonine-protein kinase PLK4 | | Authors: | Atwell, S, Burley, S.K, Houle, A, Leon, B, Pelletier, L.A, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of PLK4 kinase.
To be Published
|
|
3COM
 
 | | Crystal structure of Mst1 kinase | | Descriptor: | Serine/threonine-protein kinase 4 | | Authors: | Atwell, S, Burley, S.K, Dickey, M, Leon, B, Sauder, J.M, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Mst1 kinase.
To be Published
|
|
3PMO
 
 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4POL
 
 | | Crystal structures of thioredoxin with mesna at 2.8A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
4POK
 
 | | Crystal structures of thioredoxin with mesna at 2.5A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-25 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
4POM
 
 | | Crystal structures of thioredoxin with mesna at 1.85A resolution | | Descriptor: | 1-THIOETHANESULFONIC ACID, Thioredoxin | | Authors: | Sridhar, V, Chie-Leon, B, Badger, J, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-02-26 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BNP7787 Forms Novel Covalent
Adducts on Human Thioredoxin
and Modulates Thioredoxin
Activity
J Pharmacol Clin Toxicol, 2, 2014
|
|
4TT7
 
 | | Crystal structure of human ALK with a covalent modification | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ALK tyrosine kinase receptor | | Authors: | Badger, J, Sridhar, V, Chie-Leon, B, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-06-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel covalent modification of human anaplastic lymphoma kinase (ALK) and potentiation of crizotinib-mediated inhibition of ALK activity by BNP7787.
Onco Targets Ther, 8, 2015
|
|
4TZU
 
 | | Crystal Structure of Murine Cereblon in Complex with Pomalidomide | | Descriptor: | Protein cereblon, S-Pomalidomide, SULFATE ION, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
to be published
|
|
4MSH
 
 | | Crystal Structure of PDE10A2 with fragment ZT0143 ((2S)-4-chloro-2,3-dihydro-1,3-benzothiazol-2-amine) | | Descriptor: | 4-chloro-1,3-benzothiazol-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MRW
 
 | | Crystal structure of PDE10A2 with fragment ZT0120 (7-chloroquinolin-4-ol) | | Descriptor: | 7-chloroquinolin-4-ol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, NIenaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MRZ
 
 | | Crystal structure of PDE10A2 with fragment ZT0429 (4-methyl-3-nitropyridin-2-amine) | | Descriptor: | 4-methyl-3-nitropyridin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSA
 
 | | Crystal structure of PDE10A2 with fragment ZT0449 (5-nitro-1H-benzimidazole) | | Descriptor: | 5-nitro-1H-benzimidazole, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSC
 
 | | Crystal structure of PDE10A2 with fragment ZT1595 (2-[(quinolin-7-yloxy)methyl]quinoline) | | Descriptor: | 2-[(quinolin-7-yloxy)methyl]quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4LM3
 
 | | Crystal structure of PDE10A2 with fragment ZT464 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydro-1,4-benzodioxin-6-yl)ethanone, NICKEL (II) ION, ... | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLX
 
 | | Crystal structure of PDE10A2 with fragment ZT434 | | Descriptor: | 4,6-dimethylpyrimidin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM2
 
 | | Crystal structure of PDE10A2 with fragment ZT462 | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-6-ylmethanol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLJ
 
 | | Crystal structure of PDE10A2 with fragment ZT214 | | Descriptor: | 2H-isoindole-1,3-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM0
 
 | | Crystal structure of PDE10A2 with fragment ZT448 | | Descriptor: | 5-NITROINDAZOLE, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
