7LCD
 
 | |
6MJL
 
 | | Crystal structure of ChREBP NLS peptide bound to importin alpha. | | Descriptor: | ChREBP Peptide ASN-TYR-TRP-LYS-ARG-ARG-ILE-GLU-VAL, Importin subunit alpha-1 | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2018-09-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of importin alpha and the nuclear localization peptide of ChREBP, and small compound inhibitors of ChREBP-importin alpha interactions.
Biochem.J., 477, 2020
|
|
8SKI
 
 | |
6UQI
 
 | |
6V5K
 
 | |
6VG6
 
 | | Crystal structure of DPO4 with 8-oxoadenine (oxoA) and dGTP* | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), DNA (5'-D(*TP*CP*AP*TP*(A38)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-07 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Promutagenic bypass of 7,8-dihydro-8-oxoadenine by translesion synthesis DNA polymerase Dpo4.
Biochem.J., 477, 2020
|
|
6VGM
 
 | | Crystal structure of DPO4 with 8-oxoadenine (oxoA) and dTTP* | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), DNA (5'-D(*TP*CP*AP*TP*(A38)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Promutagenic bypass of 7,8-dihydro-8-oxoadenine by translesion synthesis DNA polymerase Dpo4.
Biochem.J., 477, 2020
|
|
6VNP
 
 | | Crystal structure of DPO4 extension past 8-oxoadenine (oxoA) and dT | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*TP*TP*CP*AP*T*(R5M)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*T)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of DPO4 extension past 8-oxoadenine (oxoA) and dT
To Be Published
|
|
6VKP
 
 | | Crystal structure of DPO4 extension past 8-oxoadenine (oxoA) and dG | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*AP*GP*AP*TP*TP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*AP*TP*(A38)P*GP*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Insights into the mismatch discrimination mechanism of Y-family DNA polymerase Dpo4.
Biochem.J., 478, 2021
|
|
8FOG
 
 | |
8FN3
 
 | |
6W5X
 
 | | Crystal structure of human polymerase eta complexed with N7-benzylguanine and dCTP* | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*(DRG)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of human polymerase eta complexed with N7-benzylguanine and dCTP*
To Be Published
|
|
6WK6
 
 | | Crystal structure of human polymerase eta complexed with Xanthine containing DNA | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*AP*TP*(3ZO)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-04-15 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase.
Biochem.J., 477, 2020
|
|
8G8H
 
 | |
8GKR
 
 | |
8GML
 
 | |
7L69
 
 | |
8G8J
 
 | |
8GBF
 
 | |
1DEP
 
 | | MEMBRANE PROTEIN, NMR, 1 STRUCTURE | | Descriptor: | T345-359 | | Authors: | Jung, H, Schnackerz, K.D. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR and circular dichroism studies of synthetic peptides derived from the third intracellular loop of the beta-adrenoceptor.
FEBS Lett., 358, 1995
|
|
4M2X
 
 | |
2KSG
 
 | |
5F74
 
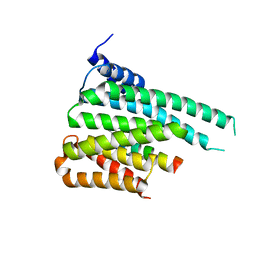 | | Crystal structure of ChREBP:14-3-3 complex bound with AMP | | Descriptor: | 14-3-3 protein beta/alpha, ADENOSINE MONOPHOSPHATE, Carbohydrate-responsive element-binding protein | | Authors: | Jung, H, Uyeda, K. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Metabolite Regulation of Nuclear Localization of Carbohydrate-response Element-binding Protein (ChREBP): ROLE OF AMP AS AN ALLOSTERIC INHIBITOR.
J.Biol.Chem., 291, 2016
|
|
6VJW
 
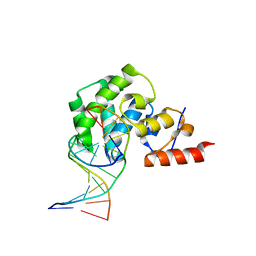 | | Crystal structure of WT hMBD4 complexed with T:G mismatch DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*CP*GP*(ORP)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4 | | Authors: | Jung, H, Lee, S. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Catalytic mechanism of the mismatch-specific DNA glycosylase methyl-CpG-binding domain 4.
Biochem.J., 477, 2020
|
|
6MQ8
 
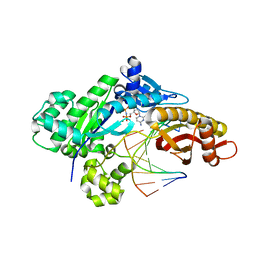 | | Binary structure of DNA polymerase eta in complex with templating hypoxanthine | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*IP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Hawkins, M.A, Jung, H, Lee, S. | | Deposit date: | 2018-10-09 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural insights into the bypass of the major deaminated purines by translesion synthesis DNA polymerase.
Biochem.J., 2020
|
|
