7WGD
 
 | |
7WGT
 
 | | X-ray structure of thermostabilized Drosophila dopamine transporter with GABA transporter1-like substitutions in the binding site, in complex with NO711. | | Descriptor: | 1-(2-{[(diphenylmethylidene)amino]oxy}ethyl)-1,2,5,6-tetrahydropyridine-3-carboxylic acid, Antibody fragment (9D5) heavy chain, Antibody fragment (9D5) light chain, ... | | Authors: | Joseph, D, Penmatsa, A. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter.
Embo J., 41, 2022
|
|
7WLW
 
 | | X-ray structure of thermostabilized Drosophila dopamine transporter with GABA transporter1-like substitutions in the binding site, in complex with SKF89976a | | Descriptor: | (3S)-1-(4,4-diphenylbut-3-enyl)piperidine-3-carboxylic acid, Antibody Fragment heavy chain, Antibody fragment light chain, ... | | Authors: | Penmatsa, A, Joseph, D. | | Deposit date: | 2022-01-13 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter.
Embo J., 41, 2022
|
|
8GNK
 
 | | CryoEM structure of cytosol-facing, substrate-bound ratGAT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Nayak, S.R, Joseph, D, Penmatsa, A. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of GABA transporter 1 reveals substrate recognition and transport mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6M0Z
 
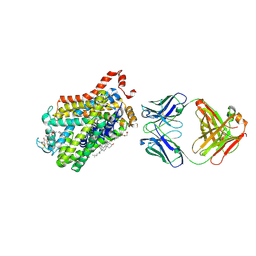 | | X-ray structure of Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in L-norepinephrine bound form | | Descriptor: | Antibody fragment (Fab) 9D5 Light chain, Antibody fragment (Fab) 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M0F
 
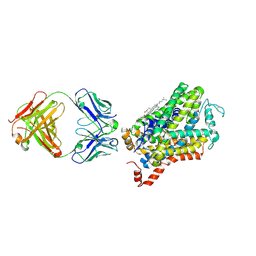 | | X-ray structure of Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) in substrate-free form | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Antibody fragment (9D5) Light chain, Antibody fragment (9D5) heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M2R
 
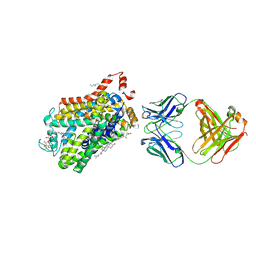 | | X-ray structure of a functional Drosophila dopamine transporter in L-norepinephrine bound form | | Descriptor: | Antibody fragment 9D5 Light chain, Antibody fragment 9D5 heavy chain, CHLORIDE ION, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-02-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M38
 
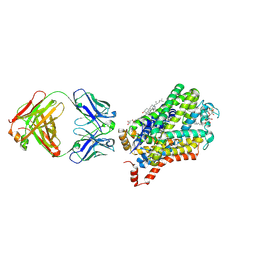 | | X-ray structure of a Drosophila dopamine transporter with subsiteB mutations (D121G/S426M) in S-duloxetine bound form | | Descriptor: | (3S)-N-methyl-3-(naphthalen-1-yloxy)-3-(thiophen-2-yl)propan-1-amine, Antibody fragment 9D5 heavy chain, Antibody fragment 9D5 light chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M3Z
 
 | | X-ray structure of a Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in milnacipran bound form | | Descriptor: | (1R,2S)-2-(aminomethyl)-N,N-diethyl-1-phenyl-cyclopropane-1-carboxamide, Antibody fragment 9D5 Light chain, Antibody fragment 9D5 heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-03-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
6M47
 
 | | X-ray structure of a Drosophila dopamine transporter with NET-like mutations (D121G/S426M/F471L) in tramadol bound form | | Descriptor: | (1S,2S)-2-[(dimethylamino)methyl]-1-(3-methoxyphenyl)cyclohexan-1-ol, Antibody fragment 9D5 light chain, Antibody fragment Heavy chain, ... | | Authors: | Shabareesh, P, Mallela, A.K, Joseph, D, Penmatsa, A. | | Deposit date: | 2020-03-05 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters.
Nat Commun, 12, 2021
|
|
5IUX
 
 | | GLIC-V135C bimane labelled X-ray structure | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, ACETATE ION, ... | | Authors: | Fourati, Z, Menny, A, Delarue, M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a pre-active conformation of a pentameric channel receptor.
Elife, 6, 2017
|
|
6IFD
 
 | |
6IFI
 
 | |
5L4H
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with 5-(2-BROMO-ETHYL)-5-ETHYL-PYRIMIDINE-2,4,6-TRIONE (brominated barbiturate) | | Descriptor: | 5-(2-bromoethyl)-5-ethyl-1,3-diazinane-2,4,6-trione, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
5L4E
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with thiopental | | Descriptor: | 5-ethyl-5-[(2R)-pentan-2-yl]-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, CHLORIDE ION, DODECANE, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
5L47
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with cyanoselenobarbital (seleniated barbiturate) | | Descriptor: | 2-[5-ethyl-2,4,6-tris(oxidanylidene)-1,3-diazinan-5-yl]ethyl selenocyanate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Reinholds Ruza, R, Fourati, Z, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
3I28
 
 | | Crystal Structure of soluble epoxide Hydrolase | | Descriptor: | 4-cyano-N-{(3S)-3-(4-fluorophenyl)-3-[4-(methylsulfonyl)phenyl]propyl}benzamide, Epoxide hydrolase 2 | | Authors: | Farrow, N.A. | | Deposit date: | 2009-06-29 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based optimization of arylamides as inhibitors of soluble epoxide hydrolase.
J.Med.Chem., 52, 2009
|
|
3I1Y
 
 | | Crystal Structure of soluble epoxide Hydrolase | | Descriptor: | Epoxide hydrolase 2, N-(3,3-diphenylpropyl)pyridine-3-carboxamide | | Authors: | Farrow, N.A. | | Deposit date: | 2009-06-28 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Structure-based optimization of arylamides as inhibitors of soluble epoxide hydrolase.
J.Med.Chem., 52, 2009
|
|
