2QRZ
 
 | | Cdc42 bound to GMP-PCP: Induced Fit by Effector is Required | | Descriptor: | Cell division control protein 42 homolog precursor, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Phillips, M.J, Calero, G, Chan, B, Cerione, R.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effector Proteins Exert an Important Influence on the Signaling-active State of the Small GTPase Cdc42.
J.Biol.Chem., 283, 2008
|
|
3PLS
 
 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
8EQ9
 
 | | Co-crystal structure of PERK with compound 11 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
8EQD
 
 | | Co-crystal structure of PERK with compound 24 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-(3-fluorophenyl)-2-hydroxyacetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
8EQE
 
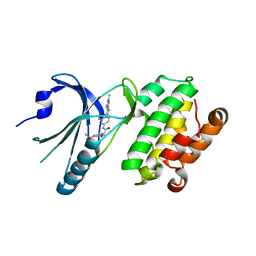 | | Co-crystal structure of PERK with compound 26 | | Descriptor: | (2R)-N-[(4M)-4-(4-amino-2,7-dimethyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-3-methylphenyl]-2-hydroxy-2-[3-(trifluoromethyl)phenyl]acetamide, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Zhu, G, Surman, M.D, Mulvihill, M.J. | | Deposit date: | 2022-10-07 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.559 Å) | | Cite: | Optimization of a Novel Mandelamide-Derived Pyrrolopyrimidine Series of PERK Inhibitors.
Pharmaceutics, 14, 2022
|
|
7MF0
 
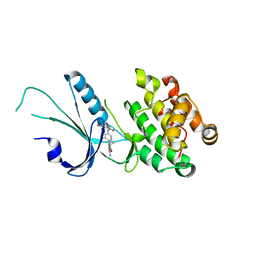 | | Co-crystal structure of PERK with inhibitor (R)-2-amino-N-cyclopropyl-5-(4-(2-(3,5-difluorophenyl)-2-hydroxyacetamido)-2-methylphenyl)nicotinamide | | Descriptor: | 2-amino-N-cyclopropyl-5-(4-{[(2R)-2-(3,5-difluorophenyl)-2-hydroxyacetyl]amino}-2-methylphenyl)pyridine-3-carboxamide, Eukaryotic translation initiation factor 2-alpha kinase 3,Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wiens, B, Koszelak-Rosenblum, M, Surman, M.D, Zhu, G, Mulvihill, M.J. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Discovery of 2-amino-3-amido-5-aryl-pyridines as highly potent, orally bioavailable, and efficacious PERK kinase inhibitors.
Bioorg.Med.Chem.Lett., 43, 2021
|
|
7NTH
 
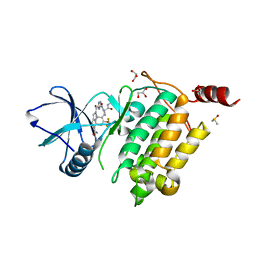 | | Structure of TAK1 in complex with compound 54 | | Descriptor: | 2-[[5-[[2-[bis(fluoranyl)methoxy]phenyl]methyl-[(2~{R})-1-(methylamino)-1-oxidanylidene-propan-2-yl]carbamoyl]-1~{H}-imidazol-2-yl]carbonyl]isoindole-5-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7NTI
 
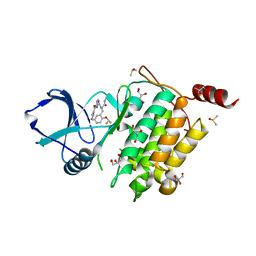 | | Structure of TAK1 in complex with compound 22 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Mitogen-activated protein kinase 7,TGF-beta-activated kinase 1 and MAP3K7-binding protein 1, ... | | Authors: | Veerman, J.J.N, Bruseker, Y.B, Damen, E, Heijne, E.H, van Bruggen, W, Hekking, K.F.W, Winkel, R, Hupp, C.D, Keefe, A.D, Liu, J, Thomson, H.A, Zhang, Y, Cuozzo, J.W, McRiner, A.J, Mulvihill, M.J, van Rijnsbergen, P, Zech, B, Renzetti, L.M, Babiss, L, Mueller, G. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of 2,4-1 H -Imidazole Carboxamides as Potent and Selective TAK1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
1SSU
 
 | | Structural and biochemical evidence for disulfide bond heterogeneity in active forms of the somatomedin B domain of human vitronectin | | Descriptor: | Vitronectin | | Authors: | Kamikubo, Y, De Guzman, R, Kroon, G, Curriden, S, Neels, J.G, Churchill, M.J, Dawson, P, Oldziej, S, Jagielska, A, Scheraga, H.A, Loskutoff, D.J, Dyson, H.J. | | Deposit date: | 2004-03-24 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin.
Biochemistry, 43, 2004
|
|
1CMQ
 
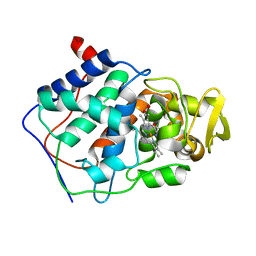 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
1CMP
 
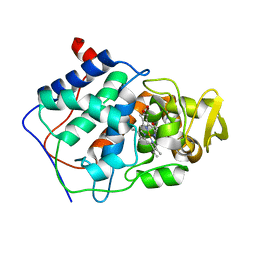 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
4ID7
 
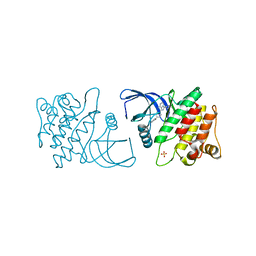 | | ACK1 kinase in complex with the inhibitor cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Descriptor: | Activated CDC42 kinase 1, SULFATE ION, cis-3-[8-amino-1-(4-phenoxyphenyl)imidazo[1,5-a]pyrazin-3-yl]cyclobutanol | | Authors: | Jin, M, Wang, J, Kleinberg, A, Kadalbajoo, M, Siu, K, Cooke, A, Bittner, M, Yao, Y, Thelemann, A, Ji, Q, Bhagwat, S, Crew, A.P, Pachter, J, Epstein, D, Mulvihill, M.J. | | Deposit date: | 2012-12-11 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, selective and orally bioavailable imidazo[1,5-a]pyrazine derived ACK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6XVF
 
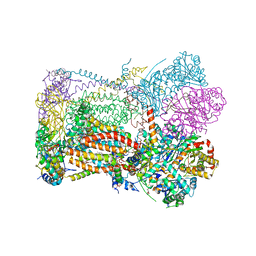 | | Crystal structure of bovine cytochrome bc1 in complex with tetrahydro-quinolone inhibitor JAG021 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-01-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Tetrahydroquinolone Eliminates Apicomplexan Parasites.
Front Cell Infect Microbiol, 10, 2020
|
|
1DT1
 
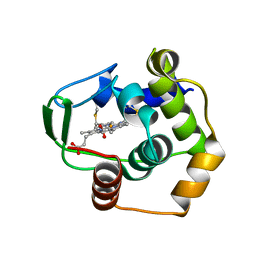 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | Deposit date: | 2000-01-10 | | Release date: | 2000-02-18 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
7SBN
 
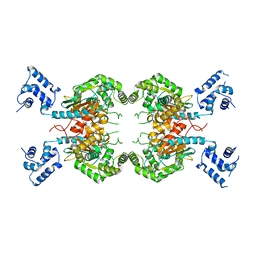 | |
7SBM
 
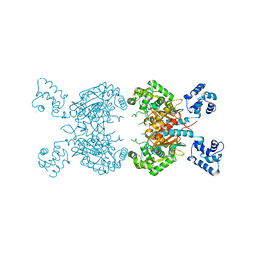 | | Human glutaminase C (Y466W) with L-Gln, open conformation | | Descriptor: | GLUTAMINE, Isoform 3 of Glutaminase kidney isoform, mitochondrial | | Authors: | Nguyen, T.-T.T, Cerione, R.A. | | Deposit date: | 2021-09-25 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-resolution structures of mitochondrial glutaminase C tetramers indicate conformational changes upon phosphate binding.
J.Biol.Chem., 298, 2022
|
|
5BXI
 
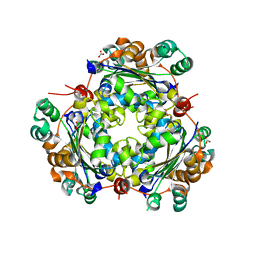 | | 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site | | Descriptor: | BICARBONATE ION, DI(HYDROXYETHYL)ETHER, Nucleoside diphosphate kinase | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-06-09 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
3D94
 
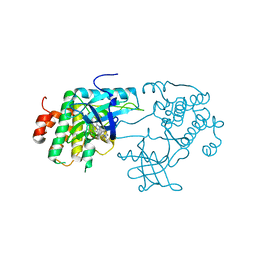 | | Crystal structure of the insulin-like growth factor-1 receptor kinase in complex with PQIP | | Descriptor: | 3-[cis-3-(4-methylpiperazin-1-yl)cyclobutyl]-1-(2-phenylquinolin-7-yl)imidazo[1,5-a]pyrazin-8-amine, CALCIUM ION, Insulin-like growth factor 1 receptor beta chain | | Authors: | Wu, J, Li, W, Miller, W.T, Hubbard, S.R. | | Deposit date: | 2008-05-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibition and activation-loop trans-phosphorylation of the IGF1 receptor
Embo J., 27, 2008
|
|
6EZM
 
 | | Imidazoleglycerol-phosphate dehydratase from Saccharomyces cerevisiae | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION, [(2R)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl]phosphonic acid | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EZJ
 
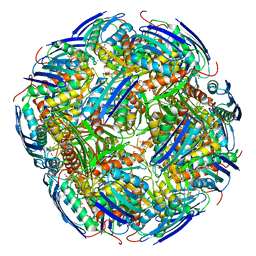 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4L3P
 
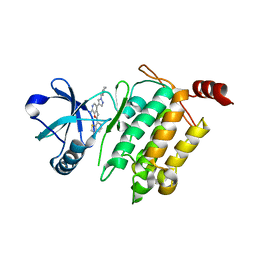 | | Crystal Structure of 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine bound to TAK1-TAB1 | | Descriptor: | 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Steinbacher, S, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4L52
 
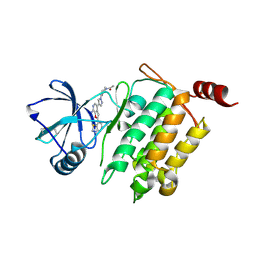 | | Crystal Structure of 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethan-1-one bound to TAK1-TAB1 | | Descriptor: | 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethanone, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | Authors: | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | Deposit date: | 2013-06-10 | | Release date: | 2013-07-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1AC4
 
 | | VARIATION IN THE STRENGTH OF A CH TO O HYDROGEN BOND IN AN ARTIFICIAL PROTEIN CAVITY (2,3,4-TRIMETHYL-1,3-THIAZOLE) | | Descriptor: | 2,3,4-TRIMETHYL-1,3-THIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-12 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variation in strength of an unconventional C-H to O hydrogen bond in an engineered protein cavity
J.Am.Chem.Soc., 119, 1997
|
|
1AEE
 
 | | SPECIFICITY OF LIGAND BINDING TO A BURIED POLAR CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE (ANILINE) | | Descriptor: | ANILINE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-24 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Artificial protein cavities as specific ligand-binding templates: characterization of an engineered heterocyclic cation-binding site that preserves the evolved specificity of the parent protein.
J.Mol.Biol., 315, 2002
|
|
1AEU
 
 | | SPECIFICITY OF LIGAND BINDING IN A POLAR CAVITY OF CYTOCHROME C PEROXIDASE (2-METHYLIMIDAZOLE) | | Descriptor: | 2-METHYLIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-25 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
