1B7F
 
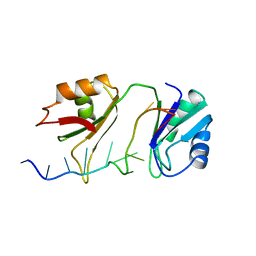 | | SXL-LETHAL PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (SXL-LETHAL PROTEIN), RNA (5'-R(P*GP*UP*UP*GP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Handa, N, Nureki, O, Kurimoto, K, Kim, I, Sakamoto, H, Shimura, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-01-23 | | Release date: | 1999-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein.
Nature, 398, 1999
|
|
1WWH
 
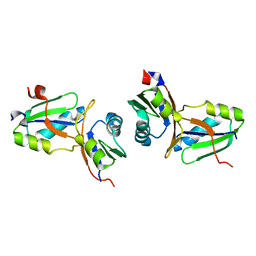 | | Crystal structure of the MPPN domain of mouse Nup35 | | Descriptor: | nucleoporin 35 | | Authors: | Handa, N, Murayama, K, Kukimoto, M, Hamana, H, Uchikubo, T, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain
J.Mol.Biol., 363, 2006
|
|
1UAN
 
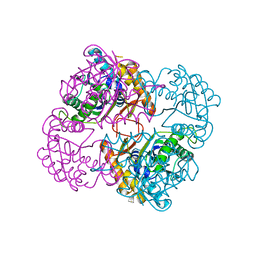 | | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TT1542 | | Authors: | Handa, N, Terada, T, Tame, J.R.H, Park, S.-Y, Kinoshita, K, Ota, M, Nakamura, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8
PROTEIN SCI., 12, 2003
|
|
1WMG
 
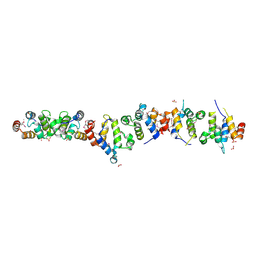 | | Crystal structure of the UNC5H2 death domain | | Descriptor: | SULFATE ION, SULFITE ION, netrin receptor Unc5h2 | | Authors: | Handa, N, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-09 | | Release date: | 2005-01-09 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the UNC5H2 death domain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
1WUB
 
 | | Crystal structure of the polyisoprenoid-binding protein, TT1927b, from Thermus thermophilus HB8 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, conserved hypothetical protein TT1927b | | Authors: | Handa, N, Idaka, M, Terada, T, Hamana, H, Ishizuka, Y, Park, S.-Y, Tame, J.R.H, Doi-Katayama, Y, Hirota, H, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-03 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a novel polyisoprenoid-binding protein from Thermus thermophilus HB8
Protein Sci., 14, 2005
|
|
2CW9
 
 | | Crystal structure of human Tim44 C-terminal domain | | Descriptor: | PENTAETHYLENE GLYCOL, translocase of inner mitochondrial membrane | | Authors: | Handa, N, Kishishita, S, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Uchikubo, T, Takemoto, C, Jin, Z, Chrzas, J, Chen, L, Liu, Z.-J, Wang, B.-C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2CWN
 
 | | Crystal structure of mouse transaldolase | | Descriptor: | Transaldolase | | Authors: | Handa, N, Arai, R, Nishino, A, Uchikubo, T, Takemoto, C, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
2YY2
 
 | | Crystal structure of the human Phosphodiesterase 9A catalytic domain complexed with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High-affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Handa, N, Shirouzu, M, Terada, T, Omori, K, Kotera, J, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2007-10-30 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human Phosphodiesterase 9A catalytic domain.
To be Published
|
|
2YXB
 
 | |
2ZMF
 
 | | Crystal structure of the C-terminal GAF domain of human phosphodiesterase 10A | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Handa, N, Kishishita, S, Mizohata, E, Omori, K, Kotera, J, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-17 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the GAF-B Domain from Human Phosphodiesterase 10A Complexed with Its Ligand, cAMP
J.Biol.Chem., 283, 2008
|
|
3AQV
 
 | | Human AMP-activated protein kinase alpha 2 subunit kinase domain (T172D) complexed with compound C | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine | | Authors: | Handa, N, Takagi, T, Saijo, S, Kishishita, S, Toyama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis for compound C inhibition of the human AMP-activated protein kinase alpha 2 subunit kinase domain
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2DB0
 
 | | Crystal structure of PH0542 | | Descriptor: | 253aa long hypothetical protein | | Authors: | Nishino, A, Handa, N, Kishishita, S, Murayama, K, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0542
To be Published
|
|
2DX0
 
 | | Crystal structure of the N-terminal SH2 domain of mouse phospholipase C-gamma 2 | | Descriptor: | Phospholipase C, gamma 2, SULFATE ION | | Authors: | Handa, N, Takagi, T, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the N-terminal SH2 domain of mouse phospholipase C-gamma 2
To be Published
|
|
4LUD
 
 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5AYF
 
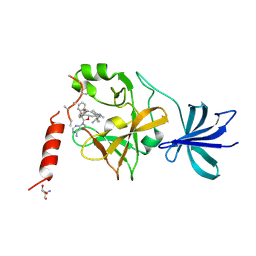 | | Crystal structure of SET7/9 in complex with cyproheptadine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dibenzo[1,2-a:2',1'-d][7]annulen-11-ylidene)-1-methyl-piperidine, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Niwa, H, Handa, N, Takemoto, Y, Ito, A, Tomabechi, Y, Umehara, T, Shirouzu, M, Yoshida, M, Yokoyama, S. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription
J.Med.Chem., 59, 2016
|
|
4ENX
 
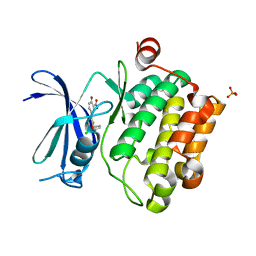 | | Crystal Structure of Pim-1 Kinase in complex with inhibitor (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-nitrobenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-nitrobenzylidene)-1,3-thiazolidin-4-one, PHOSPHATE ION, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4ENY
 
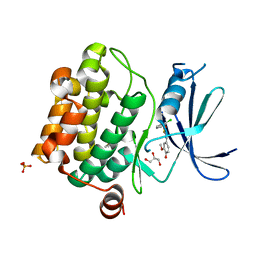 | | Crystal Structure of Pim-1 kinase in complex with (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-methoxybenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-methoxybenzylidene)-1,3-thiazolidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4LMU
 
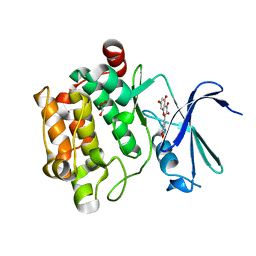 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QHE
 
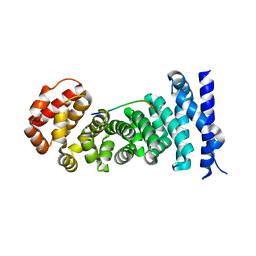 | | Crystal structure of the complex between the armadillo repeat domain of adenomatous polyposis coli and the tyrosine-rich domain of Sam68 | | Descriptor: | Adenomatous polyposis coli protein, KH domain-containing, RNA-binding, ... | | Authors: | Morishita, E.C.J, Murayama, K, Kato-Murayama, M, Ishizuku-Katsura, Y, Tomabechi, Y, Terada, T, Handa, N, Shirouzu, M, Akiyama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-25 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
4LM5
 
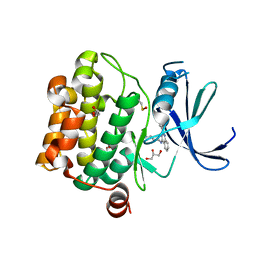 | | Crystal structure of Pim1 in complex with 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol (resulting from displacement of SKF86002) | | Descriptor: | 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUE
 
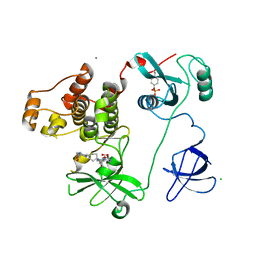 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5YVB
 
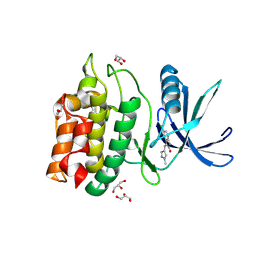 | | Structure of CaMKK2 in complex with CKI-011 | | Descriptor: | (3Z)-5-chloro-3-[(1-methyl-1H-pyrazol-4-yl)methylidene]-1,3-dihydro-2H-indol-2-one, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
5YV9
 
 | | Structure of CaMKK2 in complex with CKI-009 | | Descriptor: | 5-chloro-2-methoxy-4[(1Z)-3-(4-methoxyphenyl)-3-oxoprop-1-en-1-yl]aminobenzoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
5YV8
 
 | | Structure of CaMKK2 in complex with CKI-002 | | Descriptor: | 1-amino-4-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
