2VAN
 
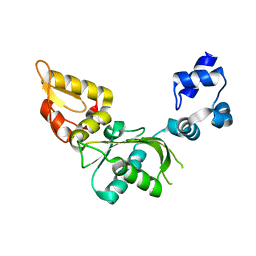 | | Nucleotidyl Transfer Mechanism of Mismatched dNTP Incorporation by DNA Polymerase b by Structural and Kinetic Analyses | | Descriptor: | DNA POLYMERSE BETA | | Authors: | Chan, H, Chou, C, Tang, K, Niebuhr, M, Tung, C, Tsai, M. | | Deposit date: | 2007-09-03 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatched Dntp Incorporation by DNA Polymerase Beta Does not Proceed Via Globally Different Conformational Pathways.
Nucleic Acids Res., 36, 2008
|
|
2K9N
 
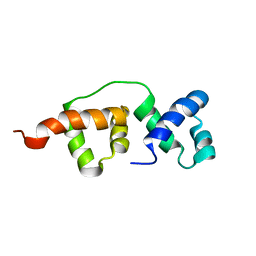 | | Solution NMR structure of the R2R3 DNA binding domain of Myb1 protein from protozoan parasite Trichomonas vaginalis | | Descriptor: | MYB24 | | Authors: | Lou, Y, Wei, S, Rajasekaran, M, Chou, C, Hsu, H, Tai, J, Chen, C. | | Deposit date: | 2008-10-19 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 37, 2009
|
|
4B8T
 
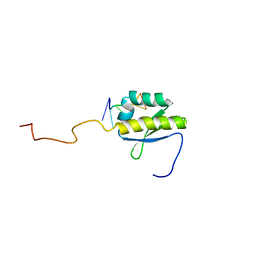 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
6MAK
 
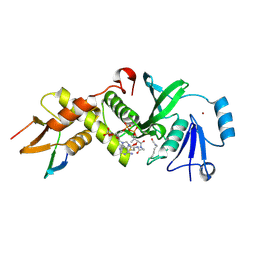 | | HBO1 is required for the maintenance of leukaemia stem cells | | Descriptor: | ACETYL COENZYME *A, BRD1 protein, Histone acetyltransferase KAT7, ... | | Authors: | Ren, B, Peat, T.S, Monahan, B, Dawson, M, Street, I. | | Deposit date: | 2018-08-27 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | HBO1 is required for the maintenance of leukaemia stem cells.
Nature, 577, 2020
|
|
6MAJ
 
 | | HBO1 is required for the maintenance of leukaemia stem cells | | Descriptor: | 4-fluoro-N'-[(3-hydroxyphenyl)sulfonyl]-5-methyl[1,1'-biphenyl]-3-carbohydrazide, BRD1 protein, GLYCEROL, ... | | Authors: | Ren, B, Peat, T.S, Monahan, B, Dawson, M, Street, I. | | Deposit date: | 2018-08-27 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.139 Å) | | Cite: | HBO1 is required for the maintenance of leukaemia stem cells.
Nature, 577, 2020
|
|
6EIH
 
 | |
4HYS
 
 | |
5KJ2
 
 | | The novel p300/CBP inhibitor A-485 uncovers a unique mechanism of action to target AR in castrate resistant prostate cancer | | Descriptor: | Histone acetyltransferase p300, N-[(4-fluorophenyl)methyl]-2-{(1R)-5-[(methylcarbamoyl)amino]-2',4'-dioxo-2,3-dihydro-3'H-spiro[indene-1,5'-[1,3]oxazolidin]-3'-yl}-N-[(2S)-1,1,1-trifluoropropan-2-yl]acetamide, SODIUM ION | | Authors: | Jakob, C.G, Qiu, W, Edalji, R.P, Sun, C. | | Deposit date: | 2016-06-17 | | Release date: | 2017-09-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours.
Nature, 550, 2017
|
|
4HYU
 
 | |
4KJU
 
 | | Crystal structure of XIAP-Bir2 with a bound benzodiazepinone inhibitor. | | Descriptor: | E3 ubiquitin-protein ligase XIAP, N-{(3S)-5-(4-aminobenzoyl)-1-[(2-methoxynaphthalen-1-yl)methyl]-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepin-3-yl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
4KJV
 
 | | Crystal structure of XIAP-Bir2 with a bound spirocyclic benzoxazepinone inhibitor. | | Descriptor: | 6-methoxy-5-({(3S)-3-[(N-methyl-L-alanyl)amino]-4-oxo-2',3,3',4,5',6'-hexahydro-5H-spiro[1,5-benzoxazepine-2,4'-pyran]-5-yl}methyl)naphthalene-2-carboxylic acid, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
1OVW
 
 | | ENDOGLUCANASE I COMPLEXED WITH NON-HYDROLYSABLE SUBSTRATE ANALOGUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-thio-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-1,4-dithio-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Sulzenbacher, G, Davies, G.J, Schulein, M. | | Deposit date: | 1996-10-17 | | Release date: | 1997-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group.
Biochemistry, 35, 1996
|
|
5MCB
 
 | | Glycogen phosphorylase in complex with chlorogenic acid. | | Descriptor: | Chlorogenic acid, Glycogen phosphorylase, muscle form, ... | | Authors: | Leonidas, D.D, Stravodimos, G.A, Kyriakis, E, Chatzileontiadou, D.S.M, Kantsadi, A.L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Affinity Crystallography Reveals the Bioactive Compounds of Industrial Juicing Byproducts of Punica granatum for Glycogen Phosphorylase.
Curr Drug Discov Technol, 15, 2018
|
|
5N22
 
 | | Structure of xEco2 acetyltransferase domain bound to K106-CoA conjugate | | Descriptor: | (2~{S})-2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]propanoic acid, GLY-ALA-LYS-LYX-ASP-GLN-TYR-PHE-LEU, XEco2 | | Authors: | Chao, W.C.H, Wade, B.O, Singleton, M.R. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis of Eco1-Mediated Cohesin Acetylation.
Sci Rep, 7, 2017
|
|
5N1W
 
 | | Structure of xEco2 acetyltransferase domain bound to K105-CoA conjugate | | Descriptor: | (2~{S})-2-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]propanoic acid, ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA,ILE-GLY-ALA-LYX-LYS-ALA, MAGNESIUM ION, ... | | Authors: | Chao, W.C.H, Wade, B.O, Singleton, M.R. | | Deposit date: | 2017-02-06 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Eco1-Mediated Cohesin Acetylation.
Sci Rep, 7, 2017
|
|
5N1U
 
 | |
1C3I
 
 | | HUMAN STROMELYSIN-1 CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, Kammlott, R.U, Crowther, R.L, Birktoft, J.J, Dunten, P, Engler, J.E. | | Deposit date: | 1999-07-27 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
1C8T
 
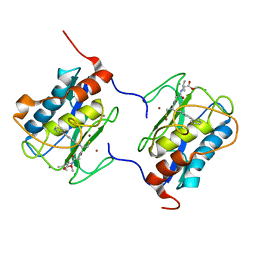 | | HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, el-Kabbani, O, Dunten, P, Crowther, R.L. | | Deposit date: | 1999-07-29 | | Release date: | 2000-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
2QIM
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, CALCIUM ION, GLYCEROL, ... | | Authors: | Fernandes, H.C, Pasternak, O, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2007-07-05 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Lupinus luteus pathogenesis-related protein as a reservoir for cytokinin.
J.Mol.Biol., 378, 2008
|
|
1IFV
 
 | |
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
1ICX
 
 | |
1QMR
 
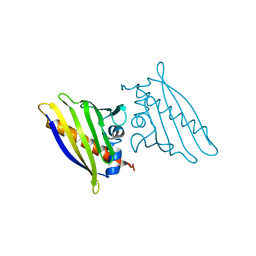 | | BIRCH POLLEN ALLERGEN BET V 1 MUTANT N28T, K32Q, E45S, P108G | | Descriptor: | MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Henriksen, A, Holm, J.O, Spangfort, M.D, Gajhede, M. | | Deposit date: | 1999-10-06 | | Release date: | 2000-10-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Allergy vaccine engineering: epitope modulation of recombinant Bet v 1 reduces IgE binding but retains protein folding pattern for induction of protective blocking-antibody responses.
J Immunol., 173, 2004
|
|
3E85
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Diphenylurea | | Descriptor: | 1,3-DIPHENYLUREA, PR10.2B, SODIUM ION | | Authors: | Fernandes, H.C, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein.
Febs J., 276, 2009
|
|
1XDF
 
 | | Crystal structure of pathogenesis-related protein LlPR-10.2A from yellow lupine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PR10.2A, SODIUM ION | | Authors: | Pasternak, O, Biesiadka, J, Dolot, R, Handschuh, L, Bujacz, G, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2004-09-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a yellow lupin pathogenesis-related PR-10 protein belonging to a novel subclass.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
