1E0F
 
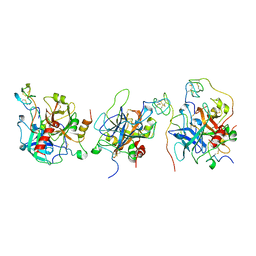 | | Crystal structure of the human alpha-thrombin-haemadin complex: an exosite II-binding inhibitor | | Descriptor: | HAEMADIN, THROMBIN | | Authors: | Richardson, J.L, Kroeger, B, Hoefken, W, Pereira, P, Huber, R, Bode, W, Fuentes-Prior, P. | | Deposit date: | 2000-03-27 | | Release date: | 2000-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Human Alpha-Thrombin-Haemadin Complex: An Exosite II-Binding Inhibitor
Embo J., 19, 2000
|
|
6EY9
 
 | |
3KD5
 
 | |
6FER
 
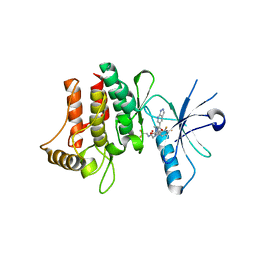 | | Crystal Structure of human DDR2 kinase in complex with 2-[4,5-difluoro-2-oxo-1'-(1H-pyrazolo[3,4-b]pyridine-5-carbonyl)spiro[indole-3,4'-piperidine]-1-yl]-N-(2,2,2-trifluoroethyl)acetamide | | Descriptor: | 2-[4,5-bis(fluoranyl)-2-oxidanylidene-1'-(1~{H}-pyrazolo[3,4-b]pyridin-5-ylcarbonyl)spiro[indole-3,4'-piperidine]-1-yl]-~{N}-[2,2,2-tris(fluoranyl)ethyl]ethanamide, Discoidin domain-containing receptor 2 | | Authors: | Stihle, M, Richter, H, Benz, J, Kuhn, B, Rudolph, M.G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | DNA-Encoded Library-Derived DDR1 Inhibitor Prevents Fibrosis and Renal Function Loss in a Genetic Mouse Model of Alport Syndrome.
Acs Chem.Biol., 14, 2019
|
|
3KD1
 
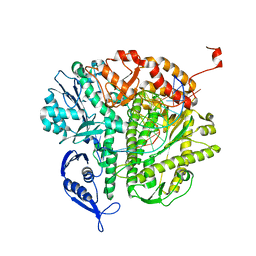 | |
6EY8
 
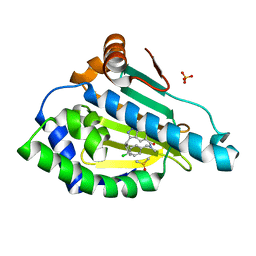 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha, SULFATE ION, ... | | Authors: | Musil, D, Lehmann, M, Buchstaller, H.-P. | | Deposit date: | 2017-11-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
6EYA
 
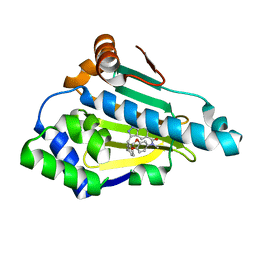 | |
6EYB
 
 | |
6F1N
 
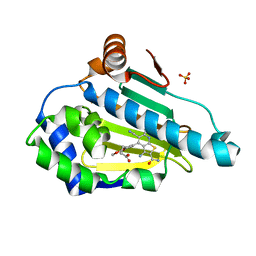 | | Estimation of relative drug-target residence times by random acceleration molecular dynamics simulation | | Descriptor: | 4-[5-[2-aminocarbonyl-3,6-bis(azanyl)-5-cyano-thieno[2,3-b]pyridin-4-yl]-2-methoxy-phenoxy]butanoic acid, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Eggenweiler, H.-M. | | Deposit date: | 2017-11-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Estimation of Drug-Target Residence Times by tau-Random Acceleration Molecular Dynamics Simulations.
J Chem Theory Comput, 14, 2018
|
|
7AOP
 
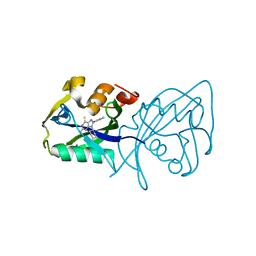 | | Structure of NUDT15 in complex with inhibitor TH8321 | | Descriptor: | 2-azanyl-9-cyclohexyl-8-(2-methoxyphenyl)-3~{H}-purine-6-thione, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
3KXM
 
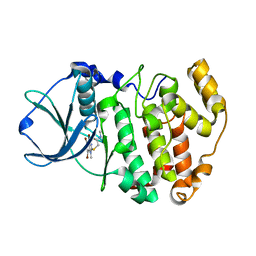 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor K74 | | Descriptor: | Casein kinase II subunit alpha, N-methyl-2-[(4,5,6,7-tetrabromo-1-methyl-1H-benzimidazol-2-yl)sulfanyl]acetamide | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
7AOM
 
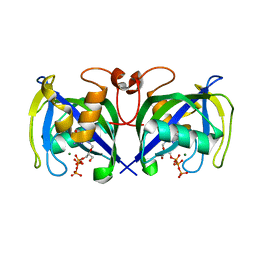 | | Structure of NUDT15 in complex with Ganciclovir triphosphate | | Descriptor: | Ganciclovir triphosphate, MAGNESIUM ION, Nucleotide triphosphate diphosphatase NUDT15 | | Authors: | Rehling, D, Zhang, S.M, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | NUDT15-mediated hydrolysis limits the efficacy of anti-HCMV drug ganciclovir.
Cell Chem Biol, 28, 2021
|
|
3KXG
 
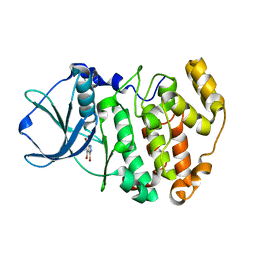 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor 3,4,5,6,7-pentabromo-1H-indazole (K64) | | Descriptor: | 3,4,5,6,7-pentabromo-1H-indazole, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
3KXH
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor (2-dymethylammino-4,5,6,7-tetrabromobenzoimidazol-1yl-acetic acid (K66) | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, [4,5,6,7-tetrabromo-2-(dimethylamino)-1H-benzimidazol-1-yl]acetic acid | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
3KXN
 
 | | Crystal structure of Z. mays CK2 kinase alpha subunit in complex with the inhibitor tetraiodobenzimidazole (K88) | | Descriptor: | 4,5,6,7-tetraiodo-1H-benzimidazole, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2009-12-03 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
6EZ8
 
 | | Human Huntingtin-HAP40 complex structure | | Descriptor: | Factor VIII intron 22 protein, Huntingtin | | Authors: | Guo, Q, Bin, H, Cheng, J, Pfeifer, G, Baumeister, W, Fernandez-Busnadiego, R, Kochanek, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-electron microscopy structure of huntingtin.
Nature, 555, 2018
|
|
4Y2H
 
 | |
4XUA
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with E11919 BAZ2-ICR analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-4-phenyl-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
4XUB
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with BAZ2-ICR chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-(1-methyl-1H-pyrazol-4-yl)-1-[2-(4-methyl-1H-1,2,3-triazol-1-yl)ethyl]-1H-imidazol-5-yl}benzonitrile, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure Enabled Design of BAZ2-ICR, A Chemical Probe Targeting the Bromodomains of BAZ2A and BAZ2B.
J.Med.Chem., 58, 2015
|
|
5XMR
 
 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and GS395 | | Descriptor: | (4~{S})-6-azanyl-3-methyl-4-[3-[4-(phenylmethyl)sulfonylphenyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-2~{H}-pyrano[2 ,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5XMS
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS498 | | Descriptor: | (4~{S})-6-azanyl-4-[3-(2-fluorophenyl)-5-(trifluoromethyl)phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5- carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5XMP
 
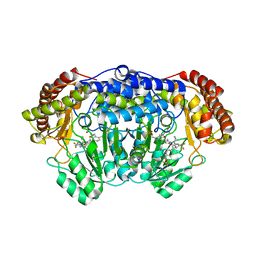 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and MF057 | | Descriptor: | 4-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-(trifluoromethyl)phenyl]-~{N}-methyl-~{N}-[2,2,2-tris(fluoranyl)ethyl]benzenesulfonamide, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5FPU
 
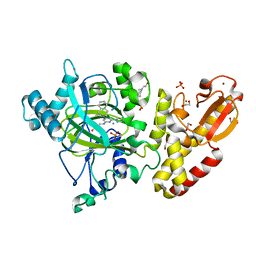 | | Crystal structure of human JARID1B in complex with GSKJ1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FWJ
 
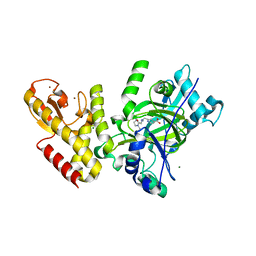 | | Crystal structure of human JARID1C in complex with KDM5-C49 | | Descriptor: | 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, HISTONE DEMETHYLASE JARID1C, MAGNESIUM ION, ... | | Authors: | Srikannathasan, V, Szykowska, A, Strain-Damerell, C, Kopec, J, Nowak, R, Gileadi, C, Johansson, C, Kupinska, K, Burgess-Brown, N.A, Shrestha, L, Dong, W, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Huber, K, Oppermann, U. | | Deposit date: | 2016-02-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5FV3
 
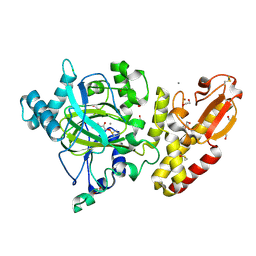 | | Crystal structure of human JARID1B construct c2 in complex with N- Oxalylglycine. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Talon, R, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2016-02-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
