5W23
 
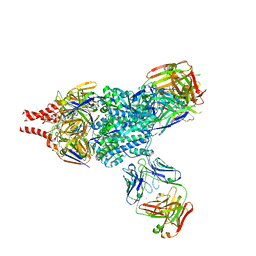 | | Crystal Structure of RSV F in complex with 5C4 Fab | | Descriptor: | 5C4 Fab heavy chain, 5C4 Fab light chain, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein.
Nat Commun, 8, 2017
|
|
5W24
 
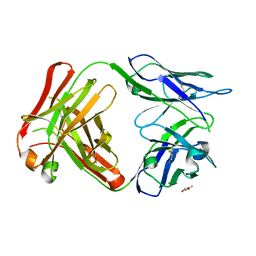 | | Crystal Structure of 5C4 Fab | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 5C4 Fab Heavy Chain, 5C4 Fab Light Chain | | Authors: | Battles, M.B, McLellan, J.S, Taher, N.M. | | Deposit date: | 2017-06-05 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein.
Nat Commun, 8, 2017
|
|
6NTE
 
 | |
7FAW
 
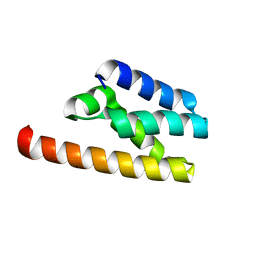 | | Structure of LW domain from Yeast | | Descriptor: | Transcription elongation factor S-II | | Authors: | Liao, S, Gao, J, Tu, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.
Int.J.Biol.Macromol., 253, 2023
|
|
8JPX
 
 | | Cryo-EM structure of PfAgo-guide DNA-target DNA complex | | Descriptor: | Excess DNA, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
6PMO
 
 | |
6POM
 
 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q0S
 
 | |
4JJJ
 
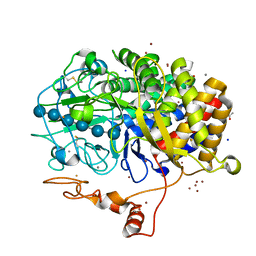 | | The structure of T. fusca GH48 D224N mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2013-03-07 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cel48A from Thermobifida fusca: structure and site directed mutagenesis of key residues.
Biotechnol.Bioeng., 111, 2014
|
|
5CVY
 
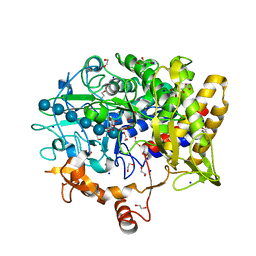 | |
6IUR
 
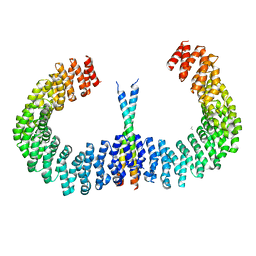 | | A phosphatase complex STRN3-PP2Aa | | Descriptor: | PP2A scaffolding subunit, PROPANE, Striatin-3 | | Authors: | Tang, Y, Zhou, Z.C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Selective Inhibition of STRN3-Containing PP2A Phosphatase Restores Hippo Tumor-Suppressor Activity in Gastric Cancer.
Cancer Cell, 38, 2020
|
|
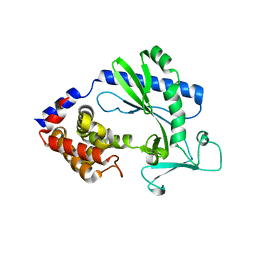
5X9C
 
 | |
5X9B
 
 | |
4O27
 
 | | Crystal structure of MST3-MO25 complex with WIF motif | | Descriptor: | 5-mer peptide from serine/threonine-protein kinase 24, ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, ... | | Authors: | Hao, Q, Feng, M, Zhou, Z.C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.185 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
4NZW
 
 | | Crystal Structure of STK25-MO25 Complex | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | Authors: | Feng, M, Hao, Q, Zhou, Z.C. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
8K3S
 
 | | Structure of PKD2-F604P complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Chen, M.Y, Su, Q, Wang, Z.F, Yu, Y. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular and structural basis of the dual regulation of the polycystin-2 ion channel by small-molecule ligands.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7Y86
 
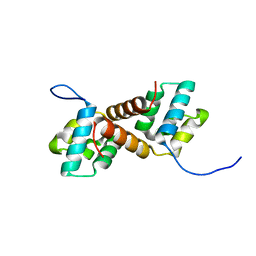 | | CcpS mutant | | Descriptor: | UPF0297 protein A7J08_00425 | | Authors: | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
7Y8Z
 
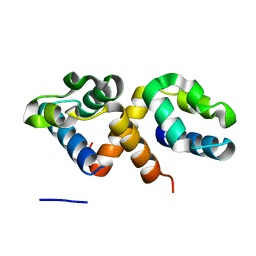 | | CcpS | | Descriptor: | UPF0297 protein A7J08_00425 | | Authors: | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
6APC
 
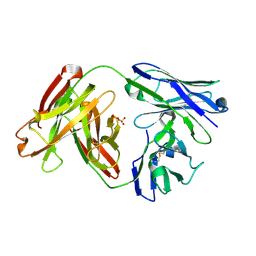 | | Crystal Structure of Infant Antibody ADI-19425 | | Descriptor: | Epididymis luminal protein 214, IGL@ protein, SULFATE ION | | Authors: | Gilman, M.S.A, McLellan, J.S. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation.
Immunity, 48, 2018
|
|
6APD
 
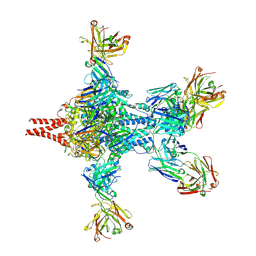 | | Crystal structure of RSV F bound by AM22 and the infant antibody ADI-19425 | | Descriptor: | AM22 Fab Heavy Chain,IGH@ protein, AM22 Fab Light Chain,Uncharacterized protein, Fusion glycoprotein F0,Envelope glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation.
Immunity, 48, 2018
|
|
6APB
 
 | |
7C24
 
 | | Glycosidase F290Y at pH8.0 | | Descriptor: | AMMONIUM ION, GLYCEROL, Isomaltose glucohydrolase | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C25
 
 | | Glycosidase Wild Type at pH8.0 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C26
 
 | | Glycosidase Wild Type at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C27
 
 | | Glycosidase F290Y at pH4.5 | | Descriptor: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
