6DWE
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and BRD0059-bound form | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-26 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
6KTK
 
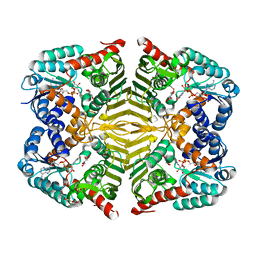 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5URC
 
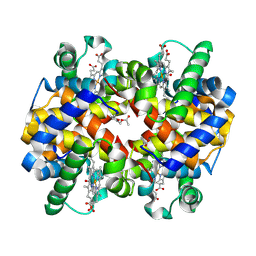 | | Design, Synthesis, Functional and Biological Evaluation of Ether and Ester Derivatives of the Antisickling Agent 5-HMF for the Treatment of Sickle Cell Disease | | Descriptor: | (5-formylfuran-2-yl)methyl acetate, 5-HYDROXYMETHYL-FURFURAL, CARBON MONOXIDE, ... | | Authors: | Pagare, P.P, Safo, R.S, Gazi, A. | | Deposit date: | 2017-02-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Ester and Ether Derivatives of Antisickling Agent 5-HMF for the Treatment of Sickle Cell Disease.
Mol. Pharm., 14, 2017
|
|
6KPC
 
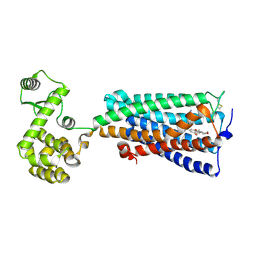 | | Crystal structure of an agonist bound GPCR | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2,Endolysin | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Wu, M, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
6DDX
 
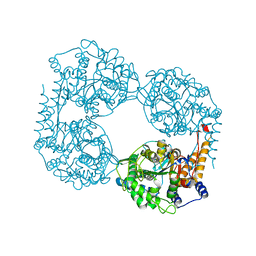 | | Crystal structure of the double mutant (D52N/L375F) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
5US1
 
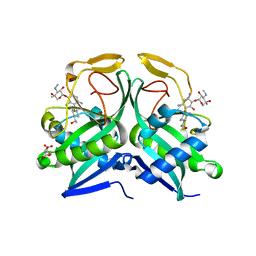 | | Crystal structure of aminoglycoside acetyltransferase AAC(2')-Ia in complex with N2'-acetylgentamicin C1A and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-(acetylamino)-6-amino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranoside, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Xu, Z, Wawrzak, Z, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
5UKA
 
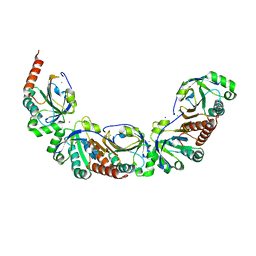 | | Salmonella typhimurium AhpC E49Q mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2017-01-20 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
6KTL
 
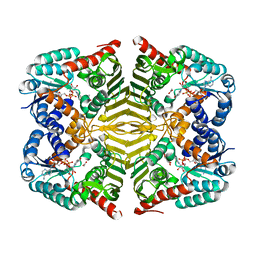 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
1AVE
 
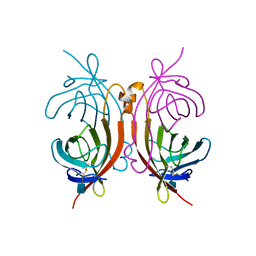 | | CRYSTAL STRUCTURE OF HEN EGG-WHITE APO-AVIDIN IN RELATION TO ITS THERMAL STABILITY PROPERTIES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AVIDIN | | Authors: | Pugliese, L, Coda, A, Malcovati, M, Bolognesi, M. | | Deposit date: | 1993-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of apo-avidin from hen egg-white.
J.Mol.Biol., 235, 1994
|
|
7CP5
 
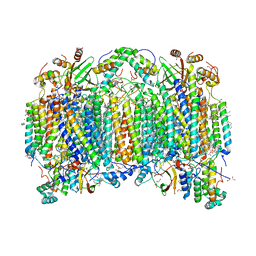 | | Bovine heart cytochrome c oxidase in a catalytic intermediate of E at 1.76 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Critical roles of the Cu B site in efficient proton pumping as revealed by crystal structures of mammalian cytochrome c oxidase catalytic intermediates.
J.Biol.Chem., 297, 2021
|
|
3H91
 
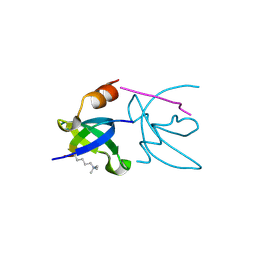 | | Crystal structure of the complex of human chromobox homolog 2 (CBX2) and H3K27 peptide | | Descriptor: | Chromobox protein homolog 2, H3K27 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
1YST
 
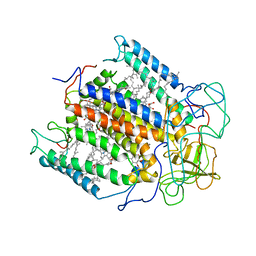 | | STRUCTURE OF THE PHOTOCHEMICAL REACTION CENTER OF A SPHEROIDENE CONTAINING PURPLE BACTERIUM, RHODOBACTER SPHAEROIDES Y, AT 3 ANGSTROMS RESOLUTION | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, MANGANESE (II) ION, ... | | Authors: | Arnoux, B, Gaucher, J.F, Ducruix, A, Reiss-Husson, F. | | Deposit date: | 1994-12-07 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the photochemical reaction centre of a spheroidene-containing purple-bacterium, Rhodobacter sphaeroides Y, at 3 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
6DD5
 
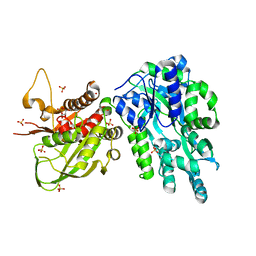 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
6DDN
 
 | |
6DJP
 
 | | Integrin alpha-v beta-8 in complex with the Fabs 8B8 and 68 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 68 heavy chain Fab, ... | | Authors: | Cormier, A, Campbell, M.G, Nishimura, S.L, Cheng, Y. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of the alpha v beta 8 integrin reveals a mechanism for stabilizing integrin extension.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DDY
 
 | | Crystal structure of the double mutant (D52N/K359Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6KXE
 
 | | The ishigamide ketosynthase/chain length factor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ketosynthase, ... | | Authors: | Du, D, Katsuyama, Y, Horiuchi, M, Fushinobu, S, Chen, A, Davis, T, Burkart, M, Ohnishi, Y. | | Deposit date: | 2019-09-10 | | Release date: | 2020-05-06 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for selectivity in a highly reducing type II polyketide synthase.
Nat.Chem.Biol., 16, 2020
|
|
1YDT
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
6KO7
 
 | | Crystal structure of the Ethidium bound RamR determined with XtaLAB Synergy | | Descriptor: | ETHIDIUM, Putative regulatory protein, SULFATE ION | | Authors: | Matsumoto, T, Nakashima, R, Yamano, A, Nishino, K. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of a structure determination method using a multidrug-resistance regulator protein as a framework.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
2JWK
 
 | |
6KPF
 
 | | Cryo-EM structure of a class A GPCR with G protein complex | | Descriptor: | 7-[(6aR,9R,10aR)-1-Hydroxy-9-(hydroxymethyl)-6,6-dimethyl-6a,7,8,9,10,10a-hexahydro-6H-benzo[c]chromen-3-yl]- 7-methyloctanenitrile, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Makriyannis, A, Shen, L, Wang, Y.X, Liu, Z.J. | | Deposit date: | 2019-08-15 | | Release date: | 2020-02-12 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-GiComplex Structures.
Cell, 180, 2020
|
|
5UTI
 
 | | Crystal Structure of TGT in complex with fragment in preQ1 pocket | | Descriptor: | DIMETHYL SULFOXIDE, L-CANAVANINE, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2017-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fragments as Novel Starting Points for tRNA-Guanine Transglycosylase Inhibitors Found by Alternative Screening Strategies.
Chemmedchem, 15, 2020
|
|
5V28
 
 | | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Dornevil, K, Liu, F, Liu, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To Be Published
|
|
5V3M
 
 | | mouseZFP568-ZnF1-11 in complex with DNA | | Descriptor: | DNA (28-MER), ZINC ION, Zinc finger protein 568 | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
1BCU
 
 | | ALPHA-THROMBIN COMPLEXED WITH HIRUGEN AND PROFLAVIN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, PROFLAVIN | | Authors: | Conti, E, Rivetti, C, Wonacott, A, Brick, P. | | Deposit date: | 1998-05-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray and spectrophotometric studies of the binding of proflavin to the S1 specificity pocket of human alpha-thrombin.
FEBS Lett., 425, 1998
|
|
