4W4R
 
 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Ko, T.P, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4XLY
 
 | | The complex structure of KS-D75C with substrate CPP | | Descriptor: | (2E)-3-methyl-5-[(1R,4aR,8aR)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4XLX
 
 | | Crystal structure of BjKS from Bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
5Y41
 
 | | Crystal Structure of LIGAND-BOUND NURR1-LBD | | Descriptor: | (13E,15S)-15-hydroxy-9-oxoprosta-10,13-dien-1-oic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sreekanth, R, Lescar, J, Yoon, H.S. | | Deposit date: | 2017-07-31 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function.
Nat.Chem.Biol., 2020
|
|
6L6R
 
 | | Crystal structure of LRP6 E1E2-SOST complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Choi, H.-J, Kim, J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Sclerostin inhibits Wnt signaling through tandem interaction with two LRP6 ectodomains.
Nat Commun, 11, 2020
|
|
1AB0
 
 | | C1G/V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN AT PH 4.5 | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C, Simpson, M, Banaszak, L, Bernlohr, D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
3UOM
 
 | | Ca2+ complex of Human skeletal calsequestrin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Sanchez, E.J, Lewis, K.M, Danna, B.R, Nissen, M.S, Kang, C.H. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | High-capacity Ca2+ Binding of Human Skeletal Calsequestrin.
J.Biol.Chem., 287, 2012
|
|
1A8Y
 
 | | CRYSTAL STRUCTURE OF CALSEQUESTRIN FROM RABBIT SKELETAL MUSCLE SARCOPLASMIC RETICULUM AT 2.4 A RESOLUTION | | Descriptor: | CALSEQUESTRIN | | Authors: | Wang, S, Trumble, W.R, Liao, H, Wesson, C.R, Dunker, A.K, Kang, C. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum.
Nat.Struct.Biol., 5, 1998
|
|
7X3T
 
 | | Cryo-EM structure of ISW1a-dinucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (343-MER), ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3X
 
 | | Cryo-EM structure of N1 nucleosome-RA | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3W
 
 | | Cryo-EM structure of ISW1-N1 nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (146-MER), ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3V
 
 | | Cryo-EM structure of IOC3-N2 nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Lifei, L, Kangjing, C, Chen, Z. | | Deposit date: | 2022-03-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5BOC
 
 | | Crystal structure of topoisomerase ParE inhibitor | | Descriptor: | 3-methyl-4-({3-[3-methyl-5-(trifluoromethyl)phenyl]-1H-pyrazol-5-yl}carbamoyl)benzoic acid, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G.Y, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
To be published
|
|
5BOD
 
 | | Crystal structure of Streptococcus pneumonia ParE inhibitor | | Descriptor: | (2R)-N-[3-(3,5-dimethylphenyl)-1H-pyrazol-5-yl]-1,4-dioxane-2-carboxamide, DNA topoisomerase 4 subunit B | | Authors: | Tan, Y.W, Chen, G, Hung, A.W, Hill, J. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Application of Fragment-based Drug Discovery against DNA GyraseB
to be published
|
|
8JVD
 
 | |
8JUC
 
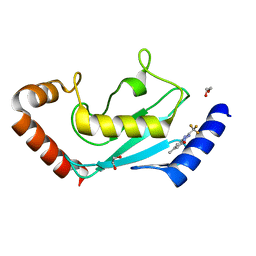 | | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening | | Descriptor: | 1,2-ETHANEDIOL, 7-methyl-2-(trifluoromethyl)-3~{H}-[1,2,4]triazolo[1,5-a]pyridin-5-one, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening.
Protein Sci., 33, 2024
|
|
6JM9
 
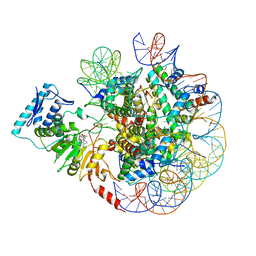 | | cryo-EM structure of DOT1L bound to unmodified nucleosome | | Descriptor: | DNA strand I, DNA strand J, Histone H2A, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
6JMA
 
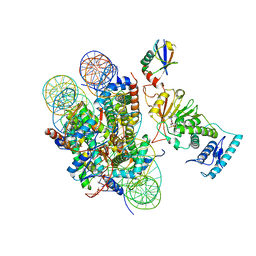 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
6L4Z
 
 | | Crystal structure of Zika NS2B-NS3 protease with compound 6 | | Descriptor: | 4-(hydroxymethyl)benzoic acid, Genome polyprotein | | Authors: | Quek, J.P. | | Deposit date: | 2019-10-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and structural characterization of small molecule fragments targeting Zika virus NS2B-NS3 protease.
Antiviral Res., 175, 2020
|
|
8HUK
 
 | | X-ray structure of human PPAR alpha ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUQ
 
 | | X-ray structure of human PPAR alpha ligand binding domain-elafibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[2,6-dimethyl-4-[(~{E})-3-(4-methylsulfanylphenyl)-3-oxidanylidene-prop-1-enyl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUL
 
 | | X-ray structure of human PPAR delta ligand binding domain-lanifibranor co-crystals obtained by co-crystallization | | Descriptor: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUN
 
 | | X-ray structure of human PPAR alpha ligand binding domain-seladelpar co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Iino, S, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUP
 
 | | X-ray structure of human PPAR gamma ligand binding domain-seladelpar-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
7F0S
 
 | |
