8ILL
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
3VJO
 
 | | Crystal structure of the wild-type EGFR kinase domain in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
3UG1
 
 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in the apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
3VJN
 
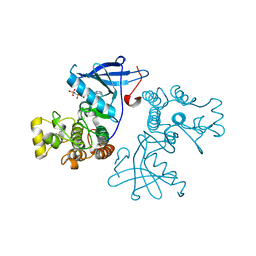 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
3UG2
 
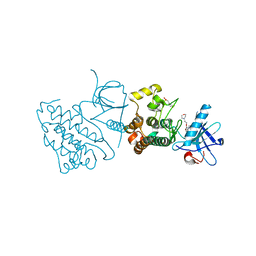 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with gefitinib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, Gefitinib | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
5B41
 
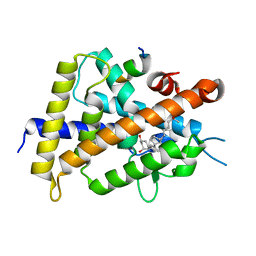 | | Crystal structure of VDR-LBD complexed with 2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Apo- and Antagonist-Binding Structures of Vitamin D Receptor Ligand-Binding Domain Revealed by Hybrid Approach Combining Small-Angle X-ray Scattering and Molecular Dynamics
J.Med.Chem., 59, 2016
|
|
2ZDY
 
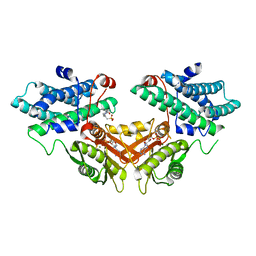 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-12-01 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2ZDX
 
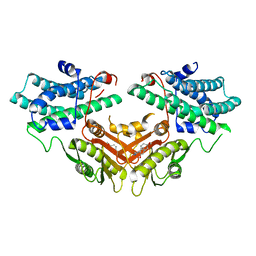 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Pyruvate dehydrogenase kinase isozyme 4 | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3P79
 
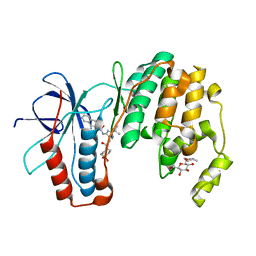 | | P38 inhibitor-bound | | Descriptor: | 1-{3-tert-butyl-1-[2-(1,1-dioxidothiomorpholin-4-yl)-2-oxoethyl]-1H-pyrazol-5-yl}-3-naphthalen-2-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5Y8U
 
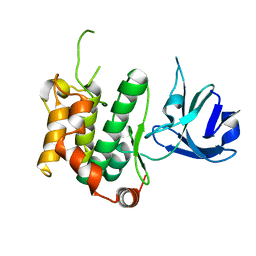 | | Crystal structure of the C276S mutant of MAP2K7 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Kinoshita, T. | | Deposit date: | 2017-08-21 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | High-resolution structure discloses the potential for allosteric regulation of mitogen-activated protein kinase kinase 7
Biochem. Biophys. Res. Commun., 493, 2017
|
|
3P78
 
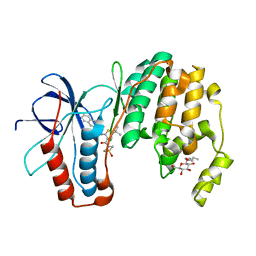 | | P38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(1,1-dioxidothiomorpholin-4-yl)carbonyl]thiophen-2-yl}-3-naphthalen-2-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P7C
 
 | | p38 inhibitor-bound | | Descriptor: | 1-[5-tert-butyl-3-({4-[2-(dimethylamino)ethyl]-5-oxo-1,4-diazepan-1-yl}carbonyl)thiophen-2-yl]-3-(2,3-dichlorophenyl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P5K
 
 | | P38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(1,1-dioxidothiomorpholin-4-yl)carbonyl]thiophen-2-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H. | | Deposit date: | 2010-10-08 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1KZK
 
 | | JE-2147-HIV Protease Complex | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Reiling, K.K, Endres, N.F, Dauber, D.S, Craik, C.S, Stroud, R.M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Anisotropic Dynamics of the JE-2147-HIV Protease Complex:
Drug Resistance and Thermodynamic Binding Mode Examined in a 1.09 A Structure
Biochemistry, 41, 2002
|
|
3P7A
 
 | | p38 inhibitor-bound | | Descriptor: | 1-[5-tert-butyl-2-(1,1-dioxidothiomorpholin-4-yl)thiophen-3-yl]-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P7B
 
 | | p38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(5-oxo-1,4-diazepan-1-yl)carbonyl]thiophen-2-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7D0I
 
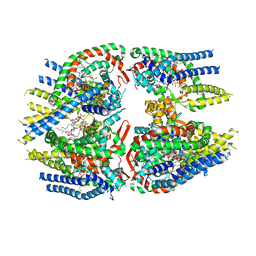 | | Cryo-EM structure of Schizosaccharomyces pombe Atg9 | | Descriptor: | Autophagy-related protein 9, Lauryl Maltose Neopentyl Glycol | | Authors: | Matoba, K, Tsutsumi, A, Kikkawa, M, Noda, N.N. | | Deposit date: | 2020-09-10 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7FG6
 
 | |
6KSV
 
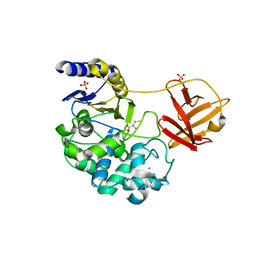 | | Crystal structure of SurE with D-Leu | | Descriptor: | Alpha/beta hydrolase, D-LEUCINE, S-(2-acetamidoethyl) (2R)-2-azanyl-4-methyl-pentanethioate, ... | | Authors: | Zhai, R, Mori, T, Abe, I. | | Deposit date: | 2019-08-26 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Heterochiral coupling in non-ribosomal peptide macrolactamization
Nat Catal, 2020
|
|
6KSU
 
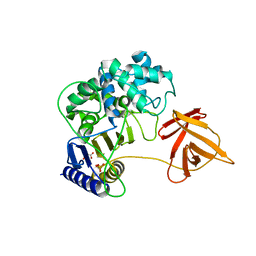 | | Crystal structure of SurE | | Descriptor: | Alpha/beta hydrolase, MALONATE ION, SULFATE ION, ... | | Authors: | Zhai, R, Mori, T, Abe, I. | | Deposit date: | 2019-08-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heterochiral coupling in non-ribosomal peptide macrolactamization
Nat Catal, 2020
|
|
7CQD
 
 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA14 tag inserted between the residues 235 and 236 | | Descriptor: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | Authors: | Tamura-Sakaguchi, R, Aruga, R, Nogi, T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Moving toward generalizable NZ-1 labeling for 3D structure determination with optimized epitope-tag insertion.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7CQC
 
 | | The NZ-1 Fab complexed with the PDZ tandem fragment of A. aeolicus S2P homolog with the PA14 tag inserted between the residues 181 and 184 | | Descriptor: | Heavy chain of antigen binding fragment, Fab of NZ-1, Light chain of antigen binding fragment, ... | | Authors: | Aruga, R, Tamura-Sakaguchi, R, Nogi, T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moving toward generalizable NZ-1 labeling for 3D structure determination with optimized epitope-tag insertion.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DBJ
 
 | | Crystal structure of human LDHB in complex with NADH, oxamate, and AXKO-0046 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase B chain, ... | | Authors: | Sogabe, S, Miwa, M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Identification of the first highly selective inhibitor of human lactate dehydrogenase B.
Sci Rep, 11, 2021
|
|
7DBK
 
 | | Crystal structure of human LDHB in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase B chain | | Authors: | Sogabe, S, Miwa, M. | | Deposit date: | 2020-10-20 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Identification of the first highly selective inhibitor of human lactate dehydrogenase B.
Sci Rep, 11, 2021
|
|
