6UH3
 
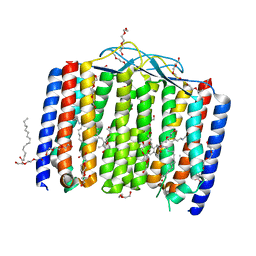 | | Crystal structure of bacterial heliorhodopsin 48C12 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heliorhodopsin, PALMITIC ACID, ... | | Authors: | Lu, Y, Zhou, X.E, Gao, X, Xia, R, Xu, Z, Wang, N, Leng, Y, Melcher, K, Xu, H.E, He, Y. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of heliorhodopsin 48C12.
Cell Res., 30, 2020
|
|
6LW3
 
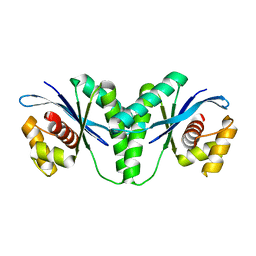 | | Crystal structure of RuvC from Pseudomonas aeruginosa | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC | | Authors: | Hu, Y, He, Y, Lin, Z. | | Deposit date: | 2020-02-07 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Biochemical and structural characterization of the Holliday junction resolvase RuvC from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 525, 2020
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
1FOU
 
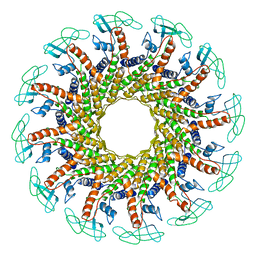 | | CONNECTOR PROTEIN FROM BACTERIOPHAGE PHI29 | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Tao, Y, Leiman, P.G, Badasso, M.O, He, Y, Jardine, P.J, Olson, N.H, Morais, M.C, Grimes, S.N, Anderson, D.L, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2000-08-28 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacteriophage phi29 DNA packaging motor.
Nature, 408, 2000
|
|
1FOQ
 
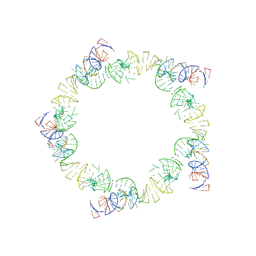 | | PENTAMERIC MODEL OF THE BACTERIOPHAGE PHI29 PROHEAD RNA | | Descriptor: | BACTERIOPHAGE PHI29 PROHEAD RNA | | Authors: | Simpson, A.A, Tao, Y, Leiman, P.G, Badasso, M.O, He, Y, Jardine, P.J, Olson, N.H, Morais, M.C, Grimes, S, Anderson, D.L, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2000-08-28 | | Release date: | 2000-12-22 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of the bacteriophage phi29 DNA packaging motor.
Nature, 408, 2000
|
|
1T4G
 
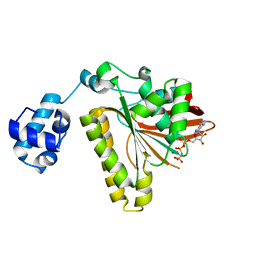 | | ATPase in complex with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, Y, He, Y, Moya, I.A, Qian, X, Luo, Y. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Archaeal Recombinase RadA; A Snapshot of Its Extended Conformation.
Mol.Cell, 15, 2004
|
|
7F7F
 
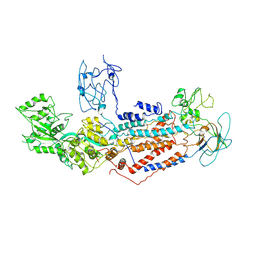 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with beryllium fluoride (resting state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-06-29 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
3FYH
 
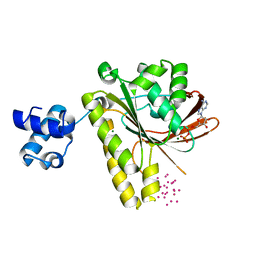 | | Recombinase in complex with ADP and metatungstate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2009-01-22 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal Rad51 homologue in complex with a metatungstate inhibitor.
Biochemistry, 48, 2009
|
|
5ZMM
 
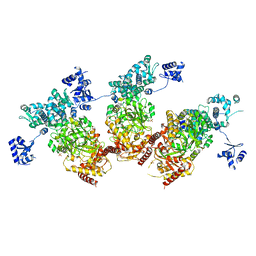 | | Structure of the Type IV phosphorothioation-dependent restriction endonuclease ScoMcrA | | Descriptor: | SULFATE ION, Uncharacterized protein McrA, ZINC ION | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
5ZMO
 
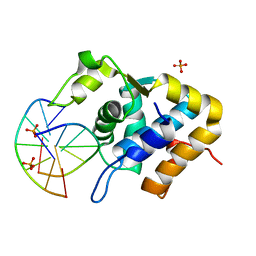 | | Sulfur binding domain of ScoMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(P*CP*CP*GP*(GS)P*CP*CP*GP*G)-3'), PHOSPHATE ION, Uncharacterized protein McrA | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
5ZMN
 
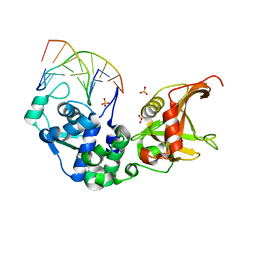 | | Sulfur binding domain and SRA domain of ScoMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*(GS)P*CP*CP*GP*GP*G)-3'), SULFATE ION, Uncharacterized protein McrA | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
6CNC
 
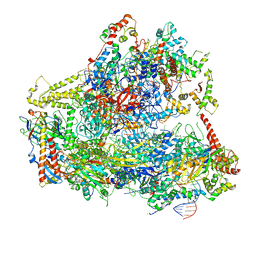 | | Yeast RNA polymerase III open complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNF
 
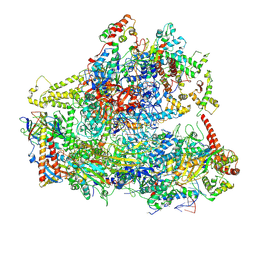 | | Yeast RNA polymerase III elongation complex | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
4IHA
 
 | | Crystal structure of rice DWARF14 (D14) in complex with a GR24 hydrolysis intermediate | | Descriptor: | (2R,3R)-2,4,4-trihydroxy-3-methylbutanal, Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH4
 
 | | Crystal structure of Arabidopsis DWARF14 orthologue, AtD14 | | Descriptor: | AT3g03990/T11I18_10 | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
6CND
 
 | | Yeast RNA polymerase III natural open complex (nOC) | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
4IH1
 
 | | Crystal structure of Karrikin Insensitive 2 (KAI2) from Arabidopsis thaliana | | Descriptor: | Hydrolase, alpha/beta fold family protein | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IH9
 
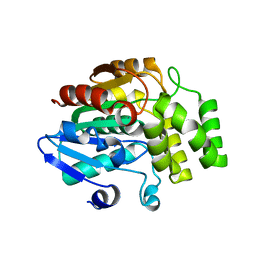 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
1JM4
 
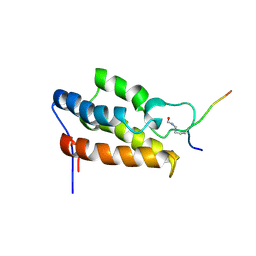 | | NMR Structure of P/CAF Bromodomain in Complex with HIV-1 Tat Peptide | | Descriptor: | HIV-1 Tat Peptide, P300/CBP-associated Factor | | Authors: | Mujtaba, S, He, Y, Zeng, L, Farooq, A, Carlson, J.E, Ott, M, Verdin, E, Zhou, M.-M. | | Deposit date: | 2001-07-17 | | Release date: | 2002-07-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain
Mol.Cell, 9, 2002
|
|
6CNB
 
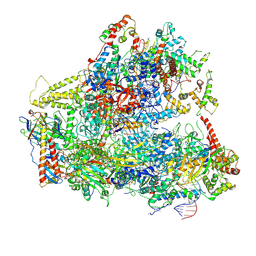 | | Yeast RNA polymerase III initial transcribing complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
1IJG
 
 | | Structure of the Bacteriophage phi29 Head-Tail Connector Protein | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Leiman, P.G, Tao, Y, He, Y, Badasso, M, Jardine, P.J, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2001-04-26 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure determination of the head-tail connector of bacteriophage phi29.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4XU5
 
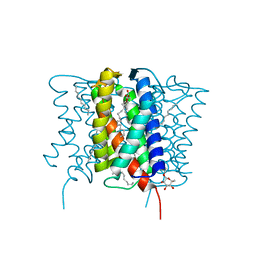 | | Crystal structure of MvINS bound to a bromine-derived 14C Diacylglycerol (DAG) at 2.1A resolution | | Descriptor: | (2S)-1-[(13-bromotridecanoyl)oxy]-3-hydroxypropan-2-yl tetradecanoate, DECANE, Uncharacterized protein, ... | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU4
 
 | | Crystal structure of a mycobacterial Insig homolog MvINS from Mycobacterium vanbaalenii at 1.9A resolution | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Uncharacterized protein, nonyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU6
 
 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
2FPL
 
 | | RadA recombinase in complex with AMP-PNP and low concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
