7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
4JBA
 
 | |
7F29
 
 | |
7X8K
 
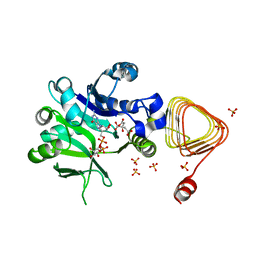 | | Arabidopsis GDP-D-mannose pyrophosphorylase (VTC1) structure (product-bound) | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, Mannose-1-phosphate guanylyltransferase 1, ... | | Authors: | Zhao, S, Zhang, C, Liu, L. | | Deposit date: | 2022-03-13 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Arabidopsis thaliana GDP-D-Mannose Pyrophosphorylase VITAMIN C DEFECTIVE 1.
Front Plant Sci, 13, 2022
|
|
7X8J
 
 | |
1TMF
 
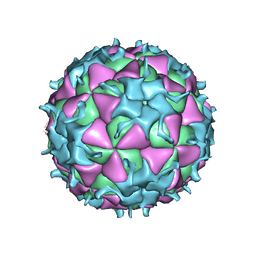 | | THREE-DIMENSIONAL STRUCTURE OF THEILER MURINE ENCEPHALOMYELITIS VIRUS (BEAN STRAIN) | | Descriptor: | THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP1), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP2), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP3), ... | | Authors: | Luo, M, Toth, K.S. | | Deposit date: | 1992-02-20 | | Release date: | 1993-10-31 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of Theiler murine encephalomyelitis virus (BeAn strain).
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
8H0N
 
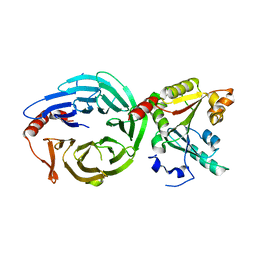 | | Crystal structure of the human METTL1-WDR4 complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 | | Authors: | Jin, X.H, Guan, Z.Y, Gong, Z, Zhang, D.L. | | Deposit date: | 2022-09-30 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into how WDR4 promotes the tRNA N7-methylguanosine methyltransferase activity of METTL1.
Cell Discov, 9, 2023
|
|
8GT9
 
 | |
8GU7
 
 | |
6KI6
 
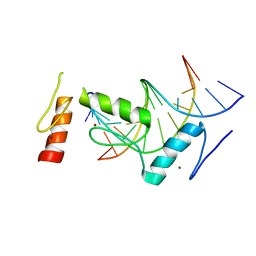 | | Crystal structure of BCL11A in complex with gamma-globin -115 HPFH region | | Descriptor: | B-cell lymphoma/leukemia 11A, DNA (5'-D(*AP*TP*AP*TP*TP*GP*GP*TP*CP*AP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*GP*AP*CP*CP*AP*AP*TP*A)-3'), ... | | Authors: | Li, F.D, Yang, Y, Shi, Y.Y. | | Deposit date: | 2019-07-17 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the recognition of gamma-globin gene promoter by BCL11A.
Cell Res., 29, 2019
|
|
7VW1
 
 | |
7VW2
 
 | |
7VW0
 
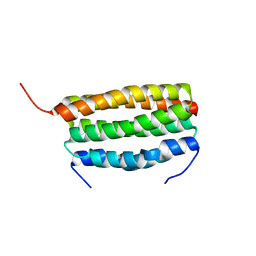 | | Structure of a dimeric periplasmic protein | | Descriptor: | DUF305 domain-containing protein | | Authors: | Yang, J, Liu, L. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.447 Å) | | Cite: | Structural basis of copper binding by a dimeric periplasmic protein forming a six-helical bundle.
J.Inorg.Biochem., 229, 2022
|
|
7VPN
 
 | |
7W5P
 
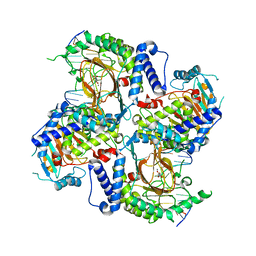 | | Crystal Structure of the dioxygenase CcTet from Coprinopsis cinereain bound to 12bp N6-methyldeoxyadenine (6mA) containing duplex DNA | | Descriptor: | CcTet, DNA, DNA (12-MER), ... | | Authors: | Mu, Y.J, Zhang, L, Zhang, L. | | Deposit date: | 2021-11-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A fungal dioxygenase CcTet serves as a eukaryotic 6mA demethylase on duplex DNA.
Nat.Chem.Biol., 18, 2022
|
|
1U3B
 
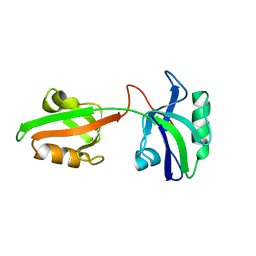 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
1U37
 
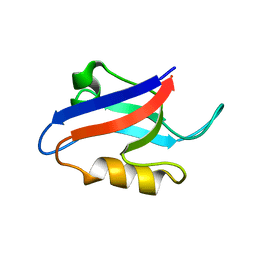 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
1U39
 
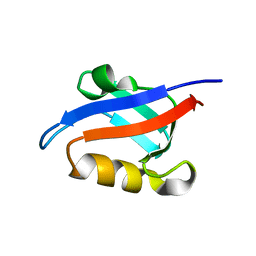 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
1U38
 
 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | PVYI, amyloid beta A4 precursor protein-binding, family A, ... | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
