2N2Q
 
 | | NMR solution structure of HsAFP1 | | Descriptor: | Defensin-like protein 1 | | Authors: | Harvey, P.J, Craik, D.J, Vriens, K. | | Deposit date: | 2015-05-11 | | Release date: | 2015-07-22 | | Last modified: | 2015-08-19 | | Method: | SOLUTION NMR | | Cite: | Synergistic Activity of the Plant Defensin HsAFP1 and Caspofungin against Candida albicans Biofilms and Planktonic Cultures.
Plos One, 10, 2015
|
|
2M2H
 
 | | Solution structure of the antimicrobial peptide [Aba3,7,12,16]BTD-2 | | Descriptor: | [Aba3,7,12,16]BTD-2 | | Authors: | Conibear, A.C, Rosengren, K, Daly, N.L, Troiera Henriques, S, Craik, D.J. | | Deposit date: | 2012-12-20 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The cyclic cystine ladder in theta-defensins is important for structure and stability, but not antibacterial activity.
J.Biol.Chem., 288, 2013
|
|
2LZI
 
 | |
2MFA
 
 | | Mambalgin-2 | | Descriptor: | Mambalgin-2 | | Authors: | Schroeder, C.I, Rash, L.D, Vila-Farres, X, Rosengren, K.J, Mobli, M, King, G.F, Alewood, P.F, Craik, D.J, Durek, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2M3I
 
 | | Characterization of a Novel Alpha4/6-Conotoxin TxIC from Conus textile that Potently Blocks alpha3beta4 Nicotinic Acetylcholine Receptors | | Descriptor: | Alpha-conotoxin | | Authors: | Luo, S, Zhangsun, D, Zhu, X, Wu, Y, Hu, Y, Christensen, S, Akcan, M, Craik, D.J, McIntosh, J.M. | | Deposit date: | 2013-01-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Novel alpha-Conotoxin TxID from Conus textile That Potently Blocks Rat alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 56, 2013
|
|
5WE3
 
 | | Solution NMR structure of PaurTx-3 | | Descriptor: | Beta-theraphotoxin-Ps1a | | Authors: | Agwa, A.J, Schroeder, C.I. | | Deposit date: | 2017-07-06 | | Release date: | 2017-09-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lengths of the C-Terminus and Interconnecting Loops Impact Stability of Spider-Derived Gating Modifier Toxins.
Toxins (Basel), 9, 2017
|
|
5JG9
 
 | |
4NE9
 
 | | PCSK9 in complex with LDLR peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low-density lipoprotein receptor, ... | | Authors: | Liu, S. | | Deposit date: | 2013-10-28 | | Release date: | 2014-09-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design and synthesis of truncated EGF-A peptides that restore LDL-R recycling in the presence of PCSK9 in vitro.
Chem.Biol., 21, 2014
|
|
6DKZ
 
 | | Racemic structure of ribifolin, an orbitide from Jatropha ribifolia | | Descriptor: | ribifolin | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-14 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
2RU2
 
 | |
3E4H
 
 | | Crystal structure of the cyclotide varv F | | Descriptor: | Varv peptide F,Varv peptide F | | Authors: | Hu, S.H. | | Deposit date: | 2008-08-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined X-ray and NMR Analysis of the Stability of the Cyclotide Cystine Knot Fold That Underpins Its Insecticidal Activity and Potential Use as a Drug Scaffold
J.Biol.Chem., 284, 2009
|
|
8FEG
 
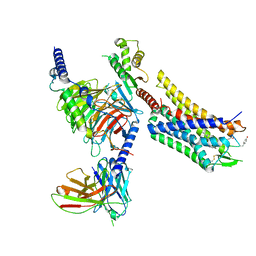 | | CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1 | | Descriptor: | ACE-TYR-ALA-DTY-THR-THR-CYS-THR-DPN-XT9, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Design and structural validation of peptide-drug conjugate ligands of the kappa-opioid receptor.
Nat Commun, 14, 2023
|
|
2BC8
 
 | | [Sec2,3,8,12]-ImI | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Armishaw, C.J. | | Deposit date: | 2005-10-18 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | {alpha}-Selenoconotoxins, a New Class of Potent {alpha}7 Neuronal Nicotinic Receptor Antagonists.
J.Biol.Chem., 281, 2006
|
|
2BC7
 
 | | Solution structure of [Sec2,8]-ImI | | Descriptor: | Alpha-conotoxin ImI | | Authors: | Armishaw, C.J. | | Deposit date: | 2005-10-18 | | Release date: | 2006-03-14 | | Last modified: | 2014-08-06 | | Method: | SOLUTION NMR | | Cite: | {alpha}-Selenoconotoxins, a New Class of Potent {alpha}7 Neuronal Nicotinic Receptor Antagonists.
J.Biol.Chem., 281, 2006
|
|
6MK4
 
 | |
6NOX
 
 | | Solution structure of SFTI-KLK5 inhibitor | | Descriptor: | SFTI-KLK5 Peptide | | Authors: | White, A.M. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Amino Acid Scanning at P5' within the Bowman-Birk Inhibitory Loop Reveals Specificity Trends for Diverse Serine Proteases.
J. Med. Chem., 62, 2019
|
|
6MK5
 
 | |
3P8F
 
 | |
3P8G
 
 | | Crystal Structure of MT-SP1 in complex with benzamidine | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, GLUTATHIONE, ... | | Authors: | Yuan, C, Huang, M, Chen, L. | | Deposit date: | 2010-10-13 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of catalytic domain of Matriptase in complex with Sunflower trypsin inhibitor-1.
Bmc Struct.Biol., 11, 2011
|
|
6CGX
 
 | | Backbone cyclised conotoxin Vc1.1 mutant - D11A, E14A | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J. | | Deposit date: | 2018-02-21 | | Release date: | 2018-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Studies Reveal the Molecular Basis for GABAB-Receptor Mediated Inhibition of High Voltage-Activated Calcium Channels by alpha-Conotoxin Vc1.1.
ACS Chem. Biol., 13, 2018
|
|
5JI4
 
 | | Solution structure of the de novo mini protein gEEHE_02 | | Descriptor: | W37 | | Authors: | Buchko, G.W, Bahl, C.D, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | Descriptor: | W35 | | Authors: | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
6OTA
 
 | |
6PI2
 
 | |
6PIN
 
 | |
