5K16
 
 | |
5K1C
 
 | | Crystal structure of the UAF1/WDR20/USP12 complex | | Descriptor: | PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Li, H, D'Andrea, A.D, Zheng, N. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Allosteric Activation of Ubiquitin-Specific Proteases by beta-Propeller Proteins UAF1 and WDR20.
Mol.Cell, 63, 2016
|
|
5K1B
 
 | |
5Y7X
 
 | | Human Peroxisome proliferator-activated receptor (PPAR) delta in complexed with a potent and selective agonist | | Descriptor: | 2-[2-methyl-4-[[4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-selenazol-5-yl]methylsulfanyl]phenoxy]ethanoic acid, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Kim, H.L, Chin, J.W, Cho, S.J, Song, J.Y, Yoon, H.S, Bae, J.H. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Highly potent and selective PPAR delta agonist reverses memory deficits in mouse models of Alzheimer's disease.
Theranostics, 14, 2024
|
|
6IJS
 
 | | Human PPARgamma ligand binding domain complexed with SB1494 | | Descriptor: | 16-mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1R,2S)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6IJR
 
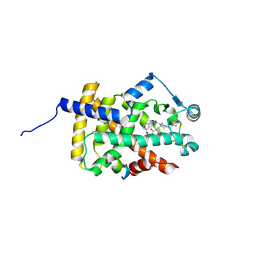 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
6PLN
 
 | |
7Y6K
 
 | |
9IXS
 
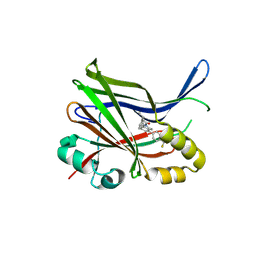 | | Crystal structure of TEAD3 YAP binding domain with compound 1 | | Descriptor: | Transcriptional enhancer factor TEF-5, ~{N}-[(1~{S})-1-phenylethyl]-5-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-isoquinoline-2-carboxamide | | Authors: | Yoo, Y. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure-based optimization of TEAD inhibitors: Exploring a novel subpocket near Glu347 for the treatment of NF2-mutant cancer.
Bioorg.Chem., 159, 2025
|
|
9IXT
 
 | | Crystal structure of TEAD3 YAP binding domain with compound 2 | | Descriptor: | Transcriptional enhancer factor TEF-5, ~{N}-[(1~{S})-4-azanyl-4-oxidanylidene-1-phenyl-butyl]-5-[4-(trifluoromethyl)phenyl]-3,4-dihydro-1~{H}-isoquinoline-2-carboxamide | | Authors: | Yoo, Y. | | Deposit date: | 2024-07-29 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of TEAD inhibitors: Exploring a novel subpocket near Glu347 for the treatment of NF2-mutant cancer.
Bioorg.Chem., 159, 2025
|
|
7XW3
 
 | |
7XW2
 
 | |
5YSS
 
 | |
5Y86
 
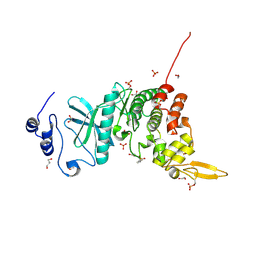 | | Crystal structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 3, ... | | Authors: | Kim, K.L, Cha, J.S, Cho, Y.S, Kim, H.Y, Chang, N.P, Cho, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
J. Mol. Biol., 430, 2018
|
|
9JRY
 
 | |
8IVU
 
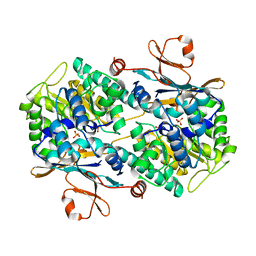 | | Crystal Structure of Human NAMPT in complex with A4276 | | Descriptor: | N-[[4-(6-methyl-1,3-benzoxazol-2-yl)phenyl]methyl]pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Kang, B.G, Cha, S.S. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09000921 Å) | | Cite: | Discovery of a novel NAMPT inhibitor that selectively targets NAPRT-deficient EMT-subtype cancer cells and alleviates chemotherapy-induced peripheral neuropathy.
Theranostics, 13, 2023
|
|
7YC5
 
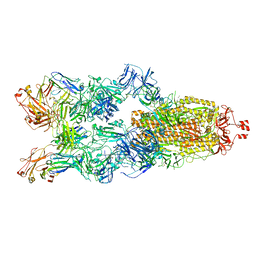 | | Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bispecific anitybody (scFv and light chain of the antibody), Heavy chain from K202.B antibody, ... | | Authors: | Yoo, Y, Cho, H.S. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-05 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Novel bispecific human antibody platform specifically targeting a fully open spike conformation potently neutralizes multiple SARS-CoV-2 variants
Antiviral Res., 212, 2023
|
|
9ERI
 
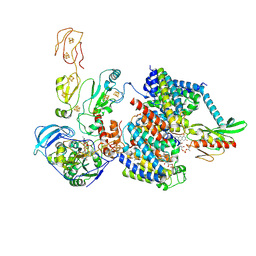 | |
9ERL
 
 | |
9ERJ
 
 | |
9ERK
 
 | |
6VC7
 
 | |
6VDF
 
 | |
8J1Q
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
9J9Z
 
 | | Cryo-EM structure of Outward state Anhydromuropeptide permease (AmpG) complex with GlcNAc-1,6-anhMurNAc | | Descriptor: | (2R)-2-[[(1R,2S,3R,4R,5R)-4-acetamido-2-[(2S,3R,4R,5S,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6,8-dioxabicyclo[3.2.1]octan-3-yl]oxy]propanoic acid, Muropeptide transporter,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, ... | | Authors: | Chang, N, Kim, U, Cho, H. | | Deposit date: | 2024-08-23 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural and functional insights of AmpG in muropeptide transport and multiple beta-lactam antibiotics resistance.
Nat Commun, 16, 2025
|
|
