2LAK
 
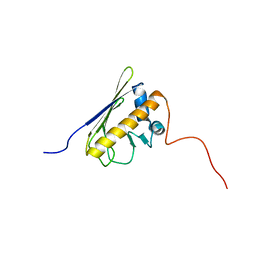 | | Solution NMR structure of the AHSA1-like protein RHE_CH02687 (1-152) from Rhizobium etli, Northeast Structural Genomics Consortium Target ReR242 | | Descriptor: | AHSA1-like protein RHE_CH02687 | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-03-16 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the AHSA1-like protein RHE_CH02687 (1-152) from Rhizobium etli, Northeast Structural Genomics Consortium Target ReR242
To be Published
|
|
2LNB
 
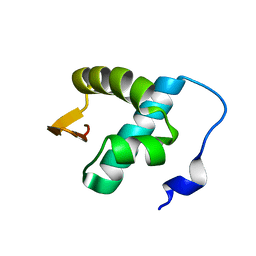 | | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A. | | Descriptor: | Z-DNA-binding protein 1 | | Authors: | Yang, Y, Ramelot, T.A, Hamilton, K, Kohan, E, Wang, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of N-terminal domain (6-74) of human ZBP1 protein, Northeast Structural Genomics Consortium Target HR8174A
To be Published
|
|
2L3A
 
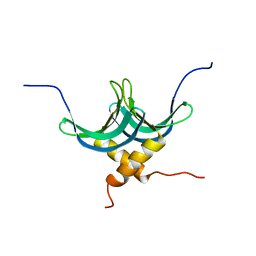 | | Solution NMR structure of homodimer protein SP_0782 (7-79) from Streptococcus pneumoniae Northeast Structural Genomics Consortium Target SpR104 . | | Descriptor: | Uncharacterized protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-10 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of hepothetical homodimer protein SP_0782 (7-79) from Streptococcus pneumoniae, Northeast Structural Genomics Consortium Target SpR104
To be Published
|
|
2LV2
 
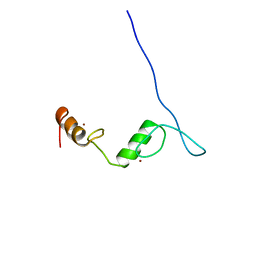 | | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B | | Descriptor: | Insulinoma-associated protein 1, ZINC ION | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural
Genomics Consortium Target HR7614B (CASP Target)
To be Published
|
|
2LX7
 
 | | Solution NMR structure of SH3 domain of growth arrest-specific protein 7 (GAS7) (fragment 1-60) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8574A | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Yang, Y, Ramelot, T.A, Dan, L, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of SH3 domain of growth arrest-specific protein 7 (GAS7) (fragment 1-60) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8574A
To be Published
|
|
2MK2
 
 | | Solution NMR structure of N-terminal domain (SH2 domain) of human Inositol polyphosphate phosphatase-like protein 1 (INPPL1) (fragment 20-117), Northeast Structural Genomics Consortium Target HR9134A | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Yang, Y, Ramelot, T.A, Janjua, H, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-01-23 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of N-terminal domain (SH2 domain) of human Inositol polyphosphate phosphatase-like protein 1 (INPPL1) (fragment 20-117), Northeast Structural Genomics Consortium Target HR9134A.
To be Published
|
|
2M47
 
 | | Solution NMR structure of the Polyketide_cyc-like protein Cgl2372 from Corynebacterium glutamicum, Northeast Structural Genomics Consortium Target CgR160 | | Descriptor: | Uncharacterized protein Cgl2373 | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Sapin, A, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Polyketide_cyc-like protein Cgl2372 from Corynebacterium glutamicum, Northeast Structural Genomics Consortium Target CgR160.
To be Published
|
|
5ZHQ
 
 | |
8QN7
 
 | | Amyloid-beta 40 type 1 filament from the leptomeninges of individual with Alzheimer's disease and cerebral amyloid angiopathy | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Yang, Y, Murzin, A.S, Peak-Chew, S.Y, Franco, C, Newell, K.L, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2023-09-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of A beta 40 filaments from the leptomeninges of individuals with Alzheimer's disease and cerebral amyloid angiopathy.
Acta Neuropathol Commun, 11, 2023
|
|
8QN6
 
 | | Amyloid-beta 40 type 2 filament from the leptomeninges of individual with Alzheimer's disease and cerebral amyloid angiopathy | | Descriptor: | Amyloid-beta A4 protein | | Authors: | Yang, Y, Murzin, A.S, Peak-Chew, S.Y, Franco, C, Newell, K.L, Ghetti, B, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2023-09-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM structures of A beta 40 filaments from the leptomeninges of individuals with Alzheimer's disease and cerebral amyloid angiopathy.
Acta Neuropathol Commun, 11, 2023
|
|
6KCZ
 
 | |
6IR5
 
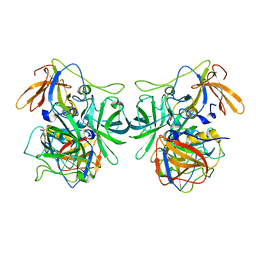 | | P domain of GII.3-TV24 | | Descriptor: | VP1 Capsid protein | | Authors: | Yang, Y. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IS5
 
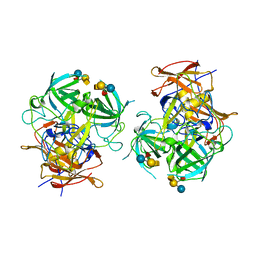 | | P domain of GII.3-TV24 with A-tetrasaccharide complex | | Descriptor: | VP1 Capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yang, Y. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6KZE
 
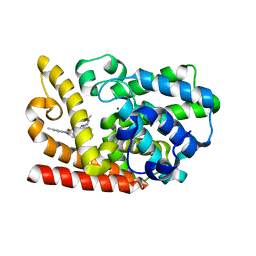 | | The crystal structue of PDE10A complexed with 4d | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.50003481 Å) | | Cite: | Novel Potent and Highly Selective Benzoimidazole-based Phosphodiesterase 10 Inhibitors with Improved Solubility and Pharmacokinetic Properties for the Treatment of Pulmonary Arterial Hypertension
To Be Published
|
|
7BPI
 
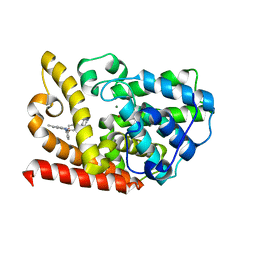 | | The crystal structue of PDE10A complexed with 14 | | Descriptor: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | Deposit date: | 2020-03-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4000864 Å) | | Cite: | Discovery of highly selective and orally available benzimidazole-based phosphodiesterase 10 inhibitors with improved solubility and pharmacokinetic properties for treatment of pulmonary arterial hypertension.
Acta Pharm Sin B, 10, 2020
|
|
7DUU
 
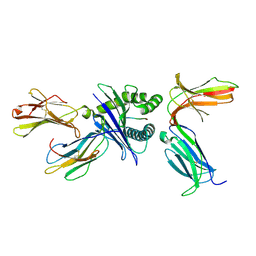 | | Crystal structure of HLA molecule with an KIR receptor | | Descriptor: | Beta-2-microglobulin, Killer cell immunoglobulin-like receptor 2DS2, LEU-ASN-PRO-SER-VAL-ALA-ALA-THR-LEU, ... | | Authors: | Yang, Y, Yin, L. | | Deposit date: | 2021-01-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Activating receptor KIR2DS2 bound to HLA-C1 reveals the novel recognition features of activating receptor.
Immunology, 165, 2022
|
|
5W4S
 
 | |
5ZTE
 
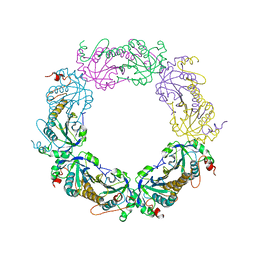 | | Crystal structure of PrxA C119S mutant from Arabidopsis thaliana | | Descriptor: | 2-Cys peroxiredoxin BAS1, chloroplastic | | Authors: | Yang, Y, Cai, W, Wang, J, Pan, W, Liu, L, Wang, M, Zhang, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabidopsis thaliana peroxiredoxin A C119S mutant.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7EYI
 
 | |
6LGK
 
 | |
6LGJ
 
 | | Crystal structure of an oxido-reductase | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Y, Lei, J, Yin, L. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an oxido-reductase
To be published
|
|
6LGM
 
 | |
6MGT
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde Decarboxylase Mutant H110A | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Daivs, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
6MGS
 
 | | Crystal structure of alpha-Amino-beta-Carboxymuconate-epsilon-Semialdehyde-Decarboxylase with Space Group of C2221 | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, COBALT (II) ION | | Authors: | Yang, Y, Davis, I, Matsui, T, Rubalcava, I, Liu, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.131 Å) | | Cite: | Quaternary structure of alpha-amino-beta-carboxymuconate-ε-semialdehyde decarboxylase (ACMSD) controls its activity.
J.Biol.Chem., 294, 2019
|
|
5NWT
 
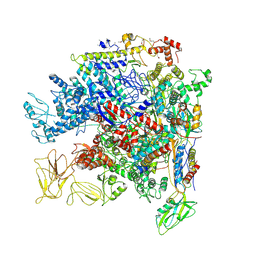 | | Crystal Structure of Escherichia coli RNA polymerase - Sigma54 Holoenzyme complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhang, X, Buck, M, Darbari, V.C, Yang, Y, Zhang, N, Lu, D, Glyde, R, Wang, Y, Winkelman, J, Gourse, R.L, Murakami, K.S. | | Deposit date: | 2017-05-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase-Sigma54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
