2PWR
 
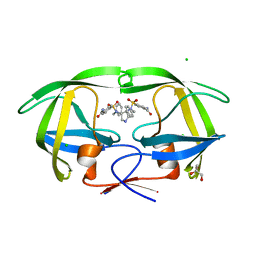 | | HIV-1 protease in complex with a carbamoyl decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-{(3S,4S)-PYRROLIDINE-3,4-DIYLBIS[(BENZYLIMINO)SULFONYL]}DIBENZAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Boettcher, B, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-05-12 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of C2-Symmetric HIV-1 Protease Inhibitors Based on a Pyrrolidine Scaffold.
J.Med.Chem., 51, 2008
|
|
3DL6
 
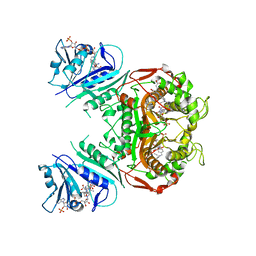 | | Crystal Structure of the A287F/S290G Active Site Mutant of TS-DHFR from Cryptosporidium hominis | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DIHYDROFOLIC ACID, ... | | Authors: | Martucci, W.E, Vargo, M.A, Anderson, K.S. | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase.
Biochemistry, 47, 2008
|
|
3DL5
 
 | | Crystal Structure of the A287F Active Site Mutant of TS-DHFR from Cryptosporidium hominis | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DIHYDROFOLIC ACID, ... | | Authors: | Vargo, M.A, Martucci, W.E, Anderson, K.S. | | Deposit date: | 2008-06-26 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Explaining an unusually fast parasitic enzyme: folate tail-binding residues dictate substrate positioning and catalysis in Cryptosporidium hominis thymidylate synthase.
Biochemistry, 47, 2008
|
|
1QQP
 
 | | FOOT-AND-MOUTH DISEASE VIRUS/ OLIGOSACCHARIDE RECEPTOR COMPLEX. | | Descriptor: | 2-O-sulfo-alpha-L-gulopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-gulopyranuronic acid, PROTEIN (GENOME POLYPROTEIN) | | Authors: | Fry, E.E, Lea, S.M, Jackson, T, Newman, J.W.I, Ellard, F.M, Blakemore, W.E, Abu-Ghazaleh, R, Samuel, A, King, A.M.Q, Stuart, D.I. | | Deposit date: | 1999-05-20 | | Release date: | 1999-06-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex.
EMBO J., 18, 1999
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
1R18
 
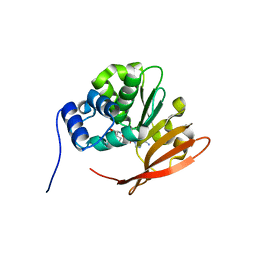 | | Drosophila protein isoaspartyl methyltransferase with S-adenosyl-L-homocysteine | | Descriptor: | Protein-L-isoaspartate(D-aspartate)-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bennett, E.J, Bjerregaard, J, Knapp, J.E, Chavous, D.A, Friedman, A.M, Royer Jr, W.E, O'Connor, C.M. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic implications from the Drosophila protein L-isoaspartyl methyltransferase structure and site-directed mutagenesis.
Biochemistry, 42, 2003
|
|
8R1T
 
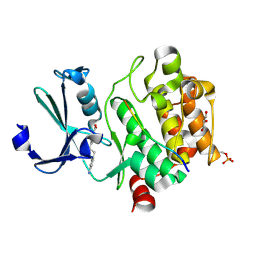 | | Pim1 in complex with 4-(4-aminophenethyl)benzoic acid and Pimtide | | Descriptor: | 4-(4-aminophenethyl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
8R1P
 
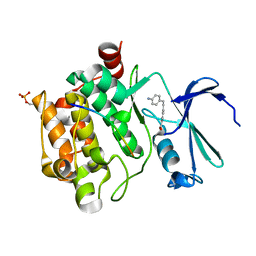 | |
8R1N
 
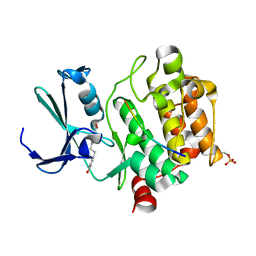 | |
8R18
 
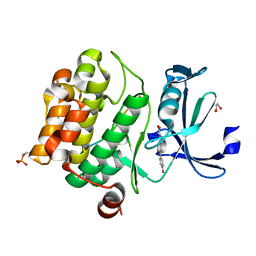 | | Pim1 in complex with (E)-4-(4-hydroxystyryl)benzoic acid and Pimtide | | Descriptor: | (E)-4-(4-hydroxystyryl)benzoic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | What doesn't fit is made to fit: Pim-1 kinase adapts to the configuration of stilbene-based inhibitors.
Arch Pharm, 357, 2024
|
|
8R10
 
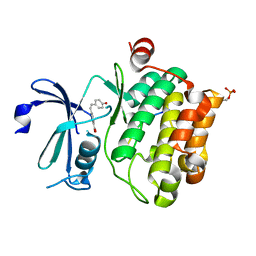 | |
1PGY
 
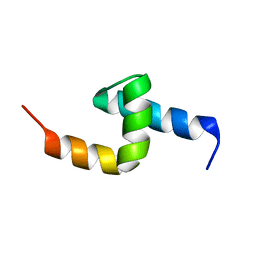 | | Solution structure of the UBA domain in Saccharomyces cerevisiae protein, Swa2p | | Descriptor: | Swa2p | | Authors: | Chim, N, Gall, W.E, Xiao, J, Harris, M.P, Graham, T.R, Krezel, A.M. | | Deposit date: | 2003-05-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-binding domain in Swa2p from Saccharomyces cerevisiae.
PROTEINS: STRUCT.,FUNCT.,GENET., 54, 2004
|
|
6CDF
 
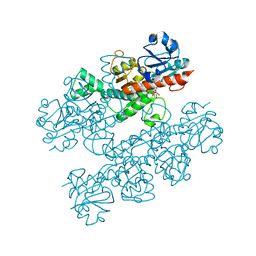 | | Human CtBP1 (28-378) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, CALCIUM ION, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
6CDR
 
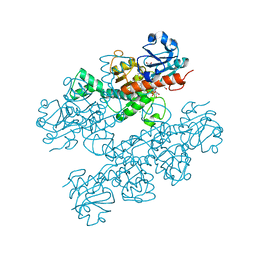 | | Human CtBP1 (28-378) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Royer, W.E, Bellesis, A.G. | | Deposit date: | 2018-02-09 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Assembly of human C-terminal binding protein (CtBP) into tetramers.
J. Biol. Chem., 293, 2018
|
|
8R0Y
 
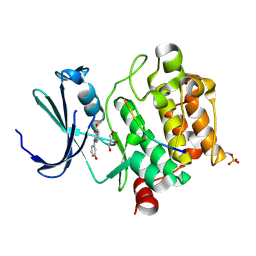 | |
8R27
 
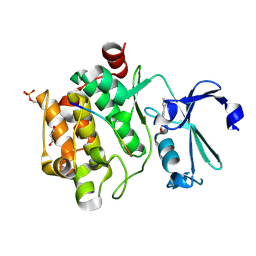 | |
8R0W
 
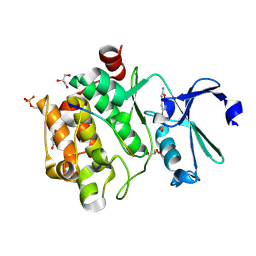 | |
8R0H
 
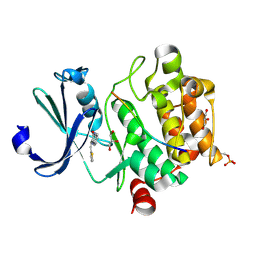 | | Pim1 in complex with (E)-3-(2-(thiophen-2-yl)vinyl)-3,4-dihydroquinoxalin-2(1H)-one and Pimtide | | Descriptor: | 3-(2-thiophen-2-ylethenyl)-1~{H}-quinoxalin-2-one, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-10-31 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Pim1 in complex with (E)-3-(2-(thiophen-2-yl)vinyl)-3,4-dihydroquinoxalin-2(1H)-one and Pimtide
To Be Published
|
|
8R1K
 
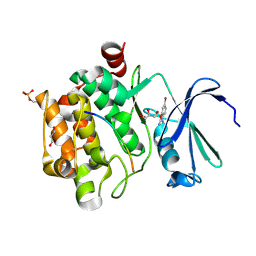 | |
8R0Q
 
 | |
8R25
 
 | |
6BQH
 
 | | Crystal structure of 5-HT2C in complex with ritanserin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, 6-(2-{4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl}ethyl)-7-methyl-5H-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ... | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
8R1W
 
 | | Pim1 in complex with 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid and Pimtide | | Descriptor: | 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2023-11-02 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pim1 in complex with 4-(5-(4-aminophenyl)-2H-1,2,3-triazol-4-yl)benzoic acid and Pimtide
To Be Published
|
|
1QRN
 
 | | CRYSTAL STRUCTURE OF HUMAN A6 TCR COMPLEXED WITH HLA-A2 BOUND TO ALTERED HTLV-1 TAX PEPTIDE P6A | | Descriptor: | BETA-2 MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, A-2 ALPHA CHAIN, ... | | Authors: | Ding, Y.H, Baker, B.M, Garboczi, D.N, Biddison, W.E, Wiley, D.C. | | Deposit date: | 1999-06-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical.
Immunity, 11, 1999
|
|
7L4E
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, ... | | Authors: | Mindrebo, J.T, Chen, A, Kim, W.E, Burkart, M.D, Noel, J.P. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases
Acs Catalysis, 11, 2021
|
|
