7BX7
 
 | |
4I4Z
 
 | | Synechocystis sp. PCC 6803 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in complex with salicylyl-CoA | | Descriptor: | BICARBONATE ION, MALONATE ION, Naphthoate synthase, ... | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4I52
 
 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
6LRQ
 
 | |
8SBL
 
 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI | | Descriptor: | Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, MHC class I antigen | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
8SBK
 
 | | Structure of HLA-A*24:02 in complex with peptide, LYLPVRVLI (ATG2A). | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, LEU-TYR-LEU-PRO-VAL-ARG-VAL-LEU-ILE, ... | | Authors: | Mallik, L, Young, M.C, Sgourakis, N.G. | | Deposit date: | 2023-04-03 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
5UQ9
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase with ((4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl)methyl dihydrogen phosphate | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating, [(4R,5R)-5-(hydroxycarbamoyl)-2,2-dimethyl-1,3-dioxolan-4-yl]methyl dihydrogen phosphate | | Authors: | Leonard, P.G. | | Deposit date: | 2017-02-07 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional Genomics Reveals Synthetic Lethality between Phosphogluconate Dehydrogenase and Oxidative Phosphorylation.
Cell Rep, 26, 2019
|
|
8JJM
 
 | |
8XUQ
 
 | |
8XUO
 
 | |
8XUV
 
 | |
7E0F
 
 | |
7VQQ
 
 | |
8EK5
 
 | | Engineered scFv 10LH bound to PHOX2B/HLA-A24:02 | | Descriptor: | 10LH single chain fragment variable (scFv), Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Garfinkle, S.E, Florio, T.J, Sgourakis, N.G. | | Deposit date: | 2022-09-20 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural principles of peptide-centric chimeric antigen receptor recognition guide therapeutic expansion.
Sci Immunol, 8, 2023
|
|
7W89
 
 | | The structure of Deinococcus radiodurans Yqgf | | Descriptor: | Putative pre-16S rRNA nuclease | | Authors: | Cheng, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.500061 Å) | | Cite: | Biochemical and Structural Study of RuvC and YqgF from Deinococcus radiodurans.
Mbio, 13, 2022
|
|
7W8D
 
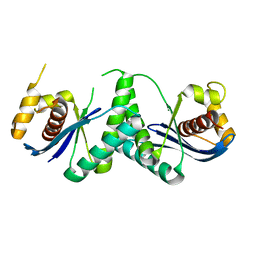 | | The structure of Deinococcus radiodurans RuvC | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, MAGNESIUM ION | | Authors: | Cheng, K. | | Deposit date: | 2021-12-07 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75016141 Å) | | Cite: | Biochemical and Structural Study of RuvC and YqgF from Deinococcus radiodurans.
Mbio, 13, 2022
|
|
7DK2
 
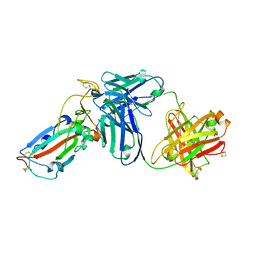 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW07 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW07 heavy chain, MW07 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M, Chen, S. | | Deposit date: | 2020-11-22 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architectural versatility of spike neutralization by a SARS-CoV-2 antibody
To Be Published
|
|
4Y2K
 
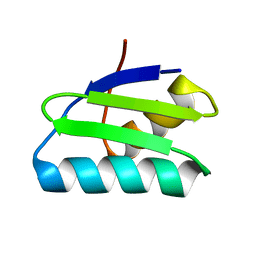 | | reduced form of apo-GolB | | Descriptor: | Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
4Y2I
 
 | | Gold ion bound to GolB | | Descriptor: | GOLD ION, Putative metal-binding transport protein | | Authors: | Wei, W, Wang, F, Ma, L, Zhao, J. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights and the Surprisingly Low Mechanical Stability of the Au-S Bond in the Gold-Specific Protein GolB
J.Am.Chem.Soc., 137, 2015
|
|
8BOB
 
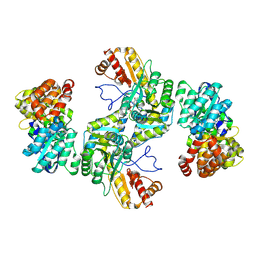 | | Structural basis for negative regulation of the maltose system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HTH-type transcriptional regulator MalT, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Chai, J, Wu, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for negative regulation of the Escherichia coli maltose system.
Nat Commun, 14, 2023
|
|
3TWJ
 
 | | Rho-associated protein kinase 1 (ROCK 1) IN COMPLEX WITH RKI1447 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3-hydroxyphenyl)methyl]-3-(4-pyridin-4-yl-1,3-thiazol-2-yl)urea, Rho-associated protein kinase 1 | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-09-21 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RKI-1447 Is a Potent Inhibitor of the Rho-Associated ROCK Kinases with Anti-Invasive and Antitumor Activities in Breast Cancer.
Cancer Res., 72, 2012
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YPG
 
 | |
7YMN
 
 | | Cryo-EM structure of in vitro PHF fibril | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Li, X, Liu, C. | | Deposit date: | 2022-07-28 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Subtle change of fibrillation condition leads to substantial alteration of recombinant Tau fibril structure.
Iscience, 25, 2022
|
|
