1DN4
 
 | | SOLVATION OF THE LEFT-HANDED HEXAMER D(5BRC-G-5BRC-G-5BRC-G) IN CRYSTALS GROWN AT TWO TEMPERATURES | | Descriptor: | DNA (5'-D(*(CBR)P*GP*(CBR)P*GP*(CBR)P*G)-3') | | Authors: | Chevrier, B, Dock, A.C, Hartmann, B, Leng, M, Moras, D, Thuong, M.T, Westhof, E. | | Deposit date: | 1986-12-01 | | Release date: | 1987-04-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures.
J.Mol.Biol., 188, 1986
|
|
2QA2
 
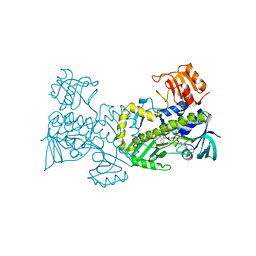 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
1GO5
 
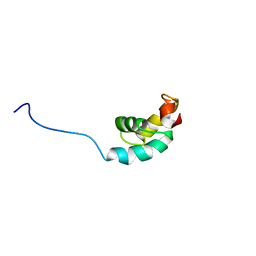 | |
2QH7
 
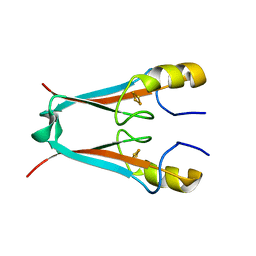 | | MitoNEET is a uniquely folded 2Fe-2S outer mitochondrial membrane protein stabilized by pioglitazone | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH-type domain 1 | | Authors: | Paddock, M.L, Wiley, S.E, Axelrod, H.L, Cohen, A.E, Roy, M, Abresch, E.C, Capraro, D, Murphy, A.N, Nechushtai, R, Dixon, J.E, Jennings, P.A. | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1GTD
 
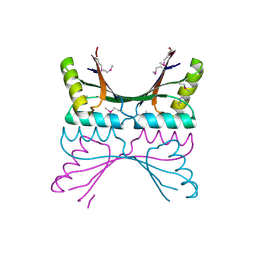 | | NORTHEAST STRUCTURAL GENOMICS CONSORTIUM (NESG ID TT50) STRUCTURE OF MTH169, THE PURS SUBUNIT OF FGAM SYNTHETASE | | Descriptor: | MTH169 | | Authors: | Batra, R, Christendat, D, Saxild, H.H, Arrowsmith, C, Tong, L. | | Deposit date: | 2002-01-14 | | Release date: | 2002-12-12 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal Structure of Mth169, a Crucial Component of Phosphoribosylformylglycinamidine Synthetase
Proteins: Struct.,Funct., Genet., 49, 2002
|
|
1GU0
 
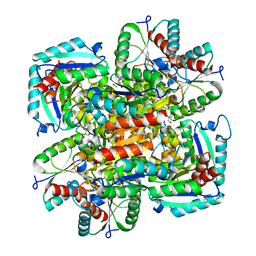 | | CRYSTAL STRUCTURE OF TYPE II DEHYDROQUINASE FROM STREPTOMYCES COELICOLOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Roszak, A.W, Krell, T, Robinson, D, Hunter, I.S, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure and Mechanism of the Type II Dehydroquinase from Streptomyces Coelicolor
Structure, 10, 2002
|
|
1GZG
 
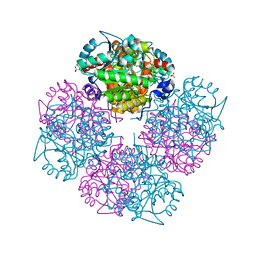 | | Complex of a Mg2-dependent porphobilinogen synthase from Pseudomonas aeruginosa (mutant D139N) with 5-fluorolevulinic acid | | Descriptor: | 5-FLUOROLEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Frere, F, Schubert, W.-D, Stauffer, F, Frankenberg, N, Neier, R, Jahn, D, Heinz, D.W. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of porphobilinogen synthase from Pseudomonas aeruginosa in complex with 5-fluorolevulinic acid suggests a double Schiff base mechanism.
J. Mol. Biol., 320, 2002
|
|
2QJ6
 
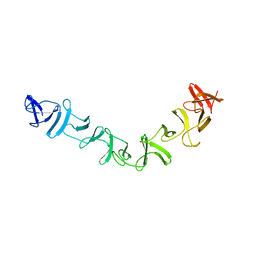 | | Crystal structure analysis of a 14 repeat C-terminal fragment of toxin TcdA in Clostridium difficile | | Descriptor: | Toxin A | | Authors: | Albesa-Jove, D, Bertrand, T, Carpenter, L, Lim, J, Brown, K.A, Fairweather, N. | | Deposit date: | 2007-07-06 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Solution and crystal structures of the cell binding domain of toxins TcdA and TcdB from Clostridium difficile
To be Published
|
|
2N4E
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34 | | Descriptor: | OR34 | | Authors: | Liu, G, Chan, K, Basanta, B, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-12-09 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34
To be Published
|
|
2N76
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414
To be Published
|
|
2N8A
 
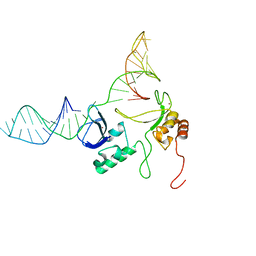 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
1H2Y
 
 | |
2MW9
 
 | |
2NPD
 
 | |
2N0T
 
 | | Structural ensemble of the enzyme cyclophilin reveals an orchestrated mode of action at atomic resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Chi, C.N, Voegeli, B, Bibow, S, Strotz, D, Orts, J, Guntert, P, Riek, R. | | Deposit date: | 2015-03-13 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Structural Ensemble for the Enzyme Cyclophilin Reveals an Orchestrated Mode of Action at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2MTL
 
 | | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109 | | Descriptor: | De novo designed protein FR55 OR109 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Ciccosanti, C, Sahdev, S, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109
To be Published
|
|
2MW5
 
 | |
1GYD
 
 | | Structure of Cellvibrio cellulosa alpha-L-arabinanase | | Descriptor: | ARABINAN ENDO-1,5-ALPHA-L-ARABINOSIDASE A | | Authors: | Nurizzo, D, Turkenburg, J.P, Charnock, S.J, Roberts, S.M, Dodson, E.J, McKie, V.A, Taylor, E.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2002-04-23 | | Release date: | 2002-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cellvibrio japonicus alpha-L-arabinanase 43A has a novel five-blade beta-propeller fold.
Nat. Struct. Biol., 9, 2002
|
|
2MWE
 
 | | NMR structure of FBP28 WW2 mutant Y438R, L453A DNDC | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Macias, M.J, Scheraga, H, Sunol, D, Todorovski, T. | | Deposit date: | 2014-11-04 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Folding kinetics of WW domains with the united residue force field for bridging microscopic motions and experimental measurements.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1GQL
 
 | | Structure of Pseudomonas cellulosa alpha-D-glucuronidase complexed with glucuronic acid and xylotriose | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-D-GLUCURONIDASE, COBALT (II) ION, ... | | Authors: | Nurizzo, D, Nagy, T, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2001-11-26 | | Release date: | 2002-09-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structural Basis for Catalysis and Specificity of the Pseudomonas Cellulosa Alpha-Glucuronidase, Glca67A
Structure, 10, 2002
|
|
1GU3
 
 | | CBM4 structure and function | | Descriptor: | ENDOGLUCANASE C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
2MOT
 
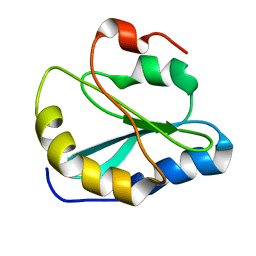 | | Backbone Structure of Actin Depolymerizing Factor (ADF) of Toxoplasma gondii Based on Prot3DNMR Approach | | Descriptor: | Actin depolymerizing factor ADF | | Authors: | Kumar, D, Raikwal, N, Raval, I, Jaiswal, N, Shukla, V, Arora, A. | | Deposit date: | 2014-05-05 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Prot3DNMR: A Simple and Swift Strategy for Backbone Structure Determination of Proteins by NMR
To be Published
|
|
2MR6
 
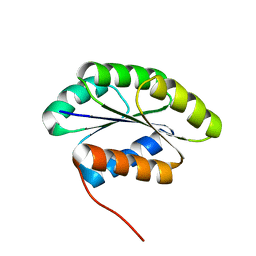 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR462 | | Descriptor: | De novo designed Protein OR462 | | Authors: | Xu, X, Nivon, L, Federizon, J.F, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR462
To be Published
|
|
1H6G
 
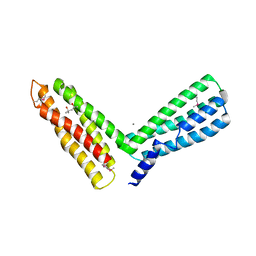 | | alpha-catenin M-domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-1 CATENIN, CALCIUM ION, ... | | Authors: | Yang, J, Dokurno, P, Tonks, N.K, Barford, D. | | Deposit date: | 2001-06-14 | | Release date: | 2001-08-07 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the M-Fragment of Alpha-Catenin: Implications for Modulation of Cell Adhesion.
Embo J., 20, 2001
|
|
1H9C
 
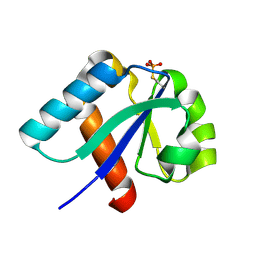 | | NMR structure of cysteinyl-phosphorylated enzyme IIB of the N,N'-diacetylchitobiose specific phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli. | | Descriptor: | PTS SYSTEM, CHITOBIOSE-SPECIFIC IIB COMPONENT | | Authors: | Ab, E, Schuurman-Wolters, G.K, Nijlant, D, Dijkstra, K, Saier, M.H, Robillard, G.T, Scheek, R.M. | | Deposit date: | 2001-03-07 | | Release date: | 2001-05-21 | | Last modified: | 2018-01-31 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Cysteinyl-Phosphorylated Enzyme Iib of the N,N'-Diacetylchitobiose Specific Phosphoenolpyruvate-Dependentphosphotransferase System of Escherichia Coli
J.Mol.Biol., 308, 2001
|
|
