4OOF
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203F mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
4FZO
 
 | |
4OOE
 
 | | M. tuberculosis 1-deoxy-d-xylulose-5-phosphate reductoisomerase W203Y mutant bound to fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Allen, C.L, Kholodar, S.A, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.826 Å) | | Cite: | Alteration of the Flexible Loop in 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase Boosts Enthalpy-Driven Inhibition by Fosmidomycin.
Biochemistry, 53, 2014
|
|
8GNJ
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
7W0N
 
 | | Cryo-EM structure of a dimeric GPCR-Gi complex with peptide | | Descriptor: | Apelin receptor early endogenous ligand, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Wu, L.J, Liu, L.E, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0L
 
 | | Cryo-EM structure of a dimeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0M
 
 | | Cryo-EM structure of a monomeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0O
 
 | | Cryo-EM structure of a monomeric GPCR-Gi complex with peptide | | Descriptor: | Apelin receptor early endogenous ligand, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0P
 
 | | Cryo-EM structure of a GPCR-Gi complex with peptide | | Descriptor: | Apelin receptor early endogenous ligand, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8HS2
 
 | | Orphan GPR20 in complex with Fab046 | | Descriptor: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HS3
 
 | | Gi bound orphan GPR20 in ligand-free state | | Descriptor: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HSC
 
 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8H85
 
 | | Trans-3/4-proline-hydroxylase H11 with 3-hydroxyl-proline | | Descriptor: | 3-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7T
 
 | | Trans-3/4-proline-hydroxylase H11 apo structure | | Descriptor: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H81
 
 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7V
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7Y
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7XE4
 
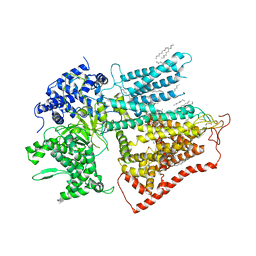 | | structure of a membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
8GNI
 
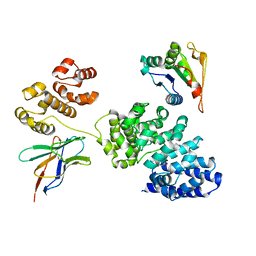 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GQ5
 
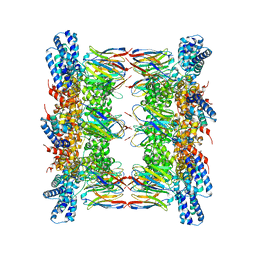 | |
4FZP
 
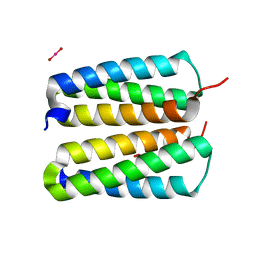 | |
2MA4
 
 | | Solution NMR Structure of yahO protein from Salmonella typhimurium, Northeast Structural Genomics Consortium (NESG) Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Eletsky, A, Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
2CPL
 
 | |
2KQP
 
 | |
2LCJ
 
 | |
