7A1K
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-((9,9-dimethyl-9H-fluoren-2-yl)methyl)-2-oxoglutarate | | Descriptor: | 3-((9,9-dimethyl-9H-fluoren-2-yl)methyl)-2-oxoglutarate, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
3B7O
 
 | | Crystal structure of the human tyrosine phosphatase SHP2 (PTPN11) with an accessible active site | | Descriptor: | D-MALATE, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Ugochukwu, E, Barr, A, Patel, A, King, O, Niesen, F, Salah, E, Savitsky, P, Pilka, E.S, Elkins, J, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2VSW
 
 | | The structure of the rhodanese domain of the human dual specificity phosphatase 16 | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 16 | | Authors: | Murray, J.W, Barr, A, Pike, A.C.W, Elkins, J, Phillips, C, Wang, J, Savitsky, P, Roos, A, Bishop, S, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Burgess-Brown, N, Pantic, N, Bray, J, von Delft, F, Gileadi, O, Knapp, S. | | Deposit date: | 2008-04-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Rhodanese Domain of the Human Dual Specifity Phosphatase 16
To be Published
|
|
2NB1
 
 | | P63/p73 hetero-tetramerisation domain | | Descriptor: | Tumor protein 63, Tumor protein p73 | | Authors: | Gebel, J, Buchner, L, Loehr, F.M, Luh, L.M, Coutandin, D, Guentert, P, Doetsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
2FCF
 
 | | The crystal structure of the 7th PDZ domain of MPDZ (MUPP-1) | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2FNE
 
 | | The crystal structure of the 13th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Berridge, G, Johansson, C, Colebrook, S, Salah, E, Burgess, N, Smee, C, Savitsky, P, Bray, J, Schoch, G, Phillips, C, Gileadi, C, Soundarajan, M, Yang, X, Elkins, J.M, Gorrec, F, Turnbull, A, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-11 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2HE4
 
 | | The crystal structure of the second PDZ domain of human NHERF-2 (SLC9A3R2) interacting with a mode 1 PDZ binding motif | | Descriptor: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2 | | Authors: | Papagrigoriou, E, Elkins, J.M, Berridge, G, Gileady, O, Colebrook, S, Gileadi, C, Salah, E, Savitsky, P, Pantic, N, Gorrec, F, Bunkoczi, G, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2PA5
 
 | | Crystal structure of human protein tyrosine phosphatase PTPN9 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THIOCYANATE ION, ... | | Authors: | Ugochukwu, E, Barr, A, Pike, A.C.W, Savitsky, P, Papagrigoriou, E, Turnbull, A, Uppenberg, J, Bunkoczi, G, Salah, E, Das, S, von Delft, F, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2P6X
 
 | | Crystal structure of human tyrosine phosphatase PTPN22 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Ugochukwu, E, Salah, E, Barr, A, Shrestha, L, Alfano, I, Burgess-Brown, N, King, O, Umeano, C, Papagrigoriou, E, Pike, A.C.W, Bunkoczi, G, Turnbull, A, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
5LF8
 
 | | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17) | | Descriptor: | ETHYL MERCURY ION, Nucleoside diphosphate-linked moiety X motif 17, PHOSPHATE ION | | Authors: | Mathea, S, Tallant, C, Salah, E, Wang, D, Velupillai, S, Nowak, R, Oerum, S, Krojer, T, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Human Nucleoside diphosphate-linked moiety X motif 17 (NUDT17)
To Be Published
|
|
5MP0
 
 | | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, m7GpppN-mRNA hydrolase | | Authors: | Mathea, S, Salah, E, Velupillai, S, Tallant, C, Pike, A.C.W, Bushell, S.R, Faust, B, Wang, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Huber, K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain
To Be Published
|
|
3G2F
 
 | | Crystal structure of the kinase domain of bone morphogenetic protein receptor type II (BMPR2) at 2.35 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bone morphogenetic protein receptor type-2, ... | | Authors: | Chaikuad, A, Thangaratnarajah, C, Roos, A.K, Filippakopoulos, P, Salah, E, Phillips, C, Keates, T, Fedorov, O, Chalk, R, Petrie, K, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-31 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural consequences of BMPR2 kinase domain mutations causing pulmonary arterial hypertension.
Sci Rep, 9, 2019
|
|
2JJD
 
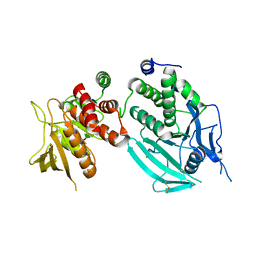 | | Protein Tyrosine Phosphatase, Receptor Type, E isoform | | Descriptor: | CHLORIDE ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE EPSILON | | Authors: | Elkins, J.M, Ugochukwu, E, Alfano, I, Barr, A.J, Bunkoczi, G, King, O.N.F, Filippakopoulos, P, Savitsky, P, Salah, E, Pike, A, Johansson, C, Das, S, Burgess-Brown, N.A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Large-Scale Structural Analysis of the Classical Human Protein Tyrosine Phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
5NKS
 
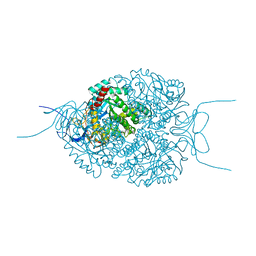 | | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4) | | Descriptor: | Dihydropyrimidinase-related protein 4 | | Authors: | Mathea, S, Elkins, J.M, Strain-Damerell, C, Salah, E, Borkowska, O, Chalk, R, Burgess-Brown, N, Pinkas, D.M, von Delft, F, Krojer, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2017-03-31 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4)
To Be Published
|
|
2PA1
 
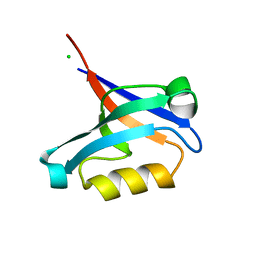 | | Structure of the PDZ domain of human PDLIM2 bound to a C-terminal extension from human beta-tropomyosin | | Descriptor: | CHLORIDE ION, PDZ and LIM domain protein 2 | | Authors: | Uppenberg, J, Shrestha, L, Elkins, J, Burgess-Brown, N, Salah, E, Bunkoczi, G, Papagrigoriou, E, Pike, A.C.W, Turnbull, A.P, Ugochukwu, E, Umeano, C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
2OOQ
 
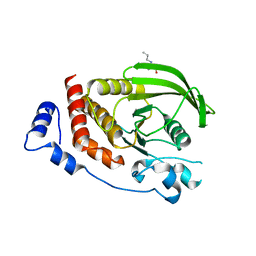 | | Crystal Structure of the Human Receptor Phosphatase PTPRT | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Receptor-type tyrosine-protein phosphatase T, ... | | Authors: | Ugochukwu, E, Alfano, I, Barr, A, Keates, T, Eswaran, J, Salah, E, Savitsky, P, Bunkoczi, G, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2QEP
 
 | | Crystal structure of the D1 domain of PTPRN2 (IA2beta) | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase N2 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Berridge, G, Burgess-Brown, N, Das, S, Fedorov, O, King, O, Niesen, F, Watt, S, Savitsky, P, Salah, E, Pike, A.C.W, Bunkoczi, G, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2RFJ
 
 | | Crystal structure of the bromo domain 1 in human bromodomain containing protein, testis specific (BRDT) | | Descriptor: | Bromodomain testis-specific protein | | Authors: | Filippakopoulos, P, Salah, E, Savitsky, P, Keates, T, Parizotto, E, Elkins, J, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2WH5
 
 | | Crystal structure of human acyl-CoA binding domain 4 complexed with stearoyl-CoA | | Descriptor: | ACYL-COA-BINDING DOMAIN-CONTAINING PROTEIN 4, COENZYME A, STEARIC ACID, ... | | Authors: | Yue, W.W, Shafqat, N, Ugochukwu, E, Savitsky, P, Johansson, C, Salah, E, Roos, A.K, Chaikuad, A, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-04-30 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Human Acyl-Coa Binding Domain 4
To be Published
|
|
2JFL
 
 | | CRYSTAL STRUCTURE OF HUMAN STE20-LIKE KINASE (DIPHOSPHORYLATED FORM) BOUND TO 5- AMINO-3-((4-(AMINOSULFONYL)PHENYL)AMINO)-N-(2,6- DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CHLORIDE ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Bunkoczi, G, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-02-02 | | Release date: | 2007-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2J51
 
 | | Crystal structure of Human STE20-like kinase bound to 5-Amino-3-((4-(aminosulfonyl)phenyl)amino) -N-(2,6-difluorophenyl)-1H-1,2,4-triazole- 1-carbothioamide | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, STE20-LIKE SERINE/THREONINE-PROTEIN KINASE, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Turnbull, A.P, Debreczeni, J.E, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-09-08 | | Release date: | 2006-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
7GH2
 
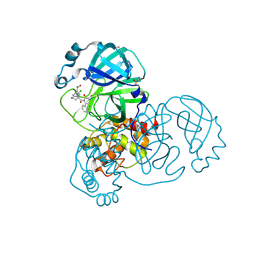 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHE
 
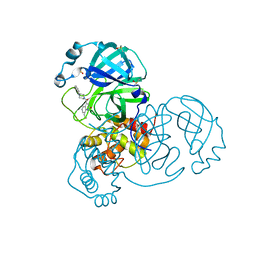 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-916a2c5a-4 (Mpro-x3303) | | Descriptor: | 3C-like proteinase, 4-(4-phenylpiperazine-1-carbonyl)quinolin-2(1H)-one, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH4
 
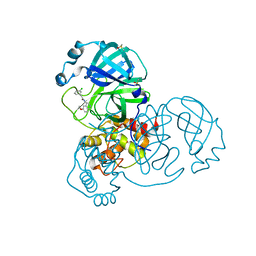 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-00c1612e-1 (Mpro-x12777) | | Descriptor: | 2-(3-chlorophenyl)-N-(6-methoxyisoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GH5
 
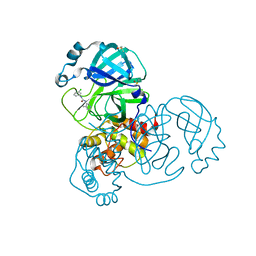 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with TRY-UNI-714-12 (Mpro-x2908) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-N'-(4-methylpyridin-3-yl)urea | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
