7SOY
 
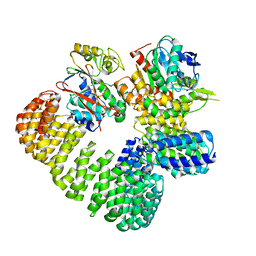 | | The structure of the PP2A-B56gamma1 holoenzyme-PME-1 complex | | Descriptor: | Isoform Gamma-1 of Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Protein phosphatase methylesterase 1, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Li, Y, Balakrishnan, V.K, Rowse, M, Novikova, I.V, Xing, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Coupling to short linear motifs creates versatile PME-1 activities in PP2A holoenzyme demethylation and inhibition.
Elife, 11, 2022
|
|
8GJZ
 
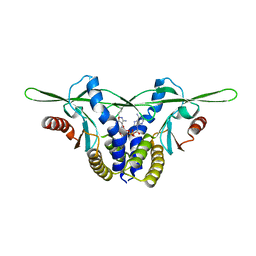 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJY
 
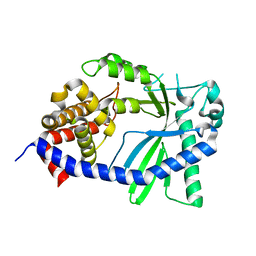 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJW
 
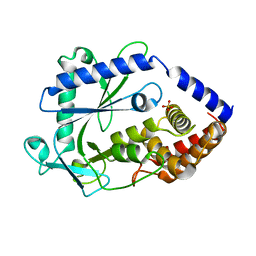 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
7E7C
 
 | |
7E7A
 
 | | Crystal structure of apo ENL YEATS domain T3 mutant | | Descriptor: | Protein ENL | | Authors: | Li, Y, Li, H. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of ENL YEATS domain T1 mutant in complex with histone H3 acetylation at K27
To Be Published
|
|
7UR2
 
 | |
7BRE
 
 | | The crystal structure of MLL2 in complex with ASH2L and RBBP5 | | Descriptor: | Histone-lysine N-methyltransferase 2B, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Zhao, L, Chen, Y. | | Deposit date: | 2020-03-28 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal Structure of MLL2 Complex Guides the Identification of a Methylation Site on P53 Catalyzed by KMT2 Family Methyltransferases.
Structure, 28, 2020
|
|
5VZT
 
 | | Crystal structure of the Skp1-FBXO31 complex | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VZU
 
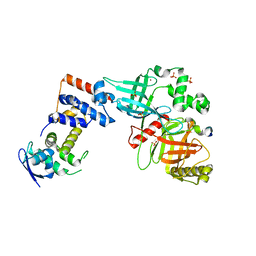 | | Crystal structure of the Skp1-FBXO31-cyclin D1 complex | | Descriptor: | Cyclin D1, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YDM
 
 | |
5YDL
 
 | |
5YDA
 
 | |
7CWW
 
 | | Crystal structure of TsrL | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, TsrE | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-08-31 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of TsrL
To Be Published
|
|
6IUS
 
 | | A higher kcat Rubisco | | Descriptor: | Ribulose-1,5-bisphosphate carboxylase/oxygenase | | Authors: | Li, Y, Cai, Z. | | Deposit date: | 2018-11-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A higher kcat Rubisco
To Be Published
|
|
6IMQ
 
 | | Crystal structure of PML B1-box multimers | | Descriptor: | CHLORIDE ION, Protein PML, ZINC ION | | Authors: | Li, Y, Ma, X, Chen, Z, Wu, H, Wang, P, Wu, W, Cheng, N, Zeng, L, Zhang, H, Cai, X, Chen, S.J, Chen, Z, Meng, G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis.
Nat Commun, 10, 2019
|
|
6N1H
 
 | | Cryo-EM structure of ASC-CARD filament | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6N1I
 
 | | Cryo-EM structure of NLRC4-CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Li, Y, Fu, T, Wu, H. | | Deposit date: | 2018-11-08 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4TMP
 
 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
4WSI
 
 | | Crystal Structure of PALS1/Crb complex | | Descriptor: | MAGUK p55 subfamily member 5, Protein crumbs | | Authors: | Wei, Z, Li, Y, Zhang, M. | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5J9S
 
 | | ENL YEATS in complex with histone H3 acetylation at K27 | | Descriptor: | H3K27ac peptide from Histone H3.1, Protein ENL, SULFATE ION | | Authors: | Li, Y, Li, H. | | Deposit date: | 2016-04-11 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Nature, 543, 2017
|
|
1FV1
 
 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
7F3B
 
 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
1SMF
 
 | | Studies on an artificial trypsin inhibitor peptide derived from the mung bean inhibitor | | Descriptor: | BOWMAN-BIRK TYPE TRYPSIN INHIBITOR, CALCIUM ION, TRYPSIN | | Authors: | Huang, Q, Li, Y, Zhang, S, Liu, S, Tang, Y, Qi, C. | | Deposit date: | 1992-10-24 | | Release date: | 1994-07-31 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies on an artificial trypsin inhibitor peptide derived from the mung bean trypsin inhibitor: chemical synthesis, refolding, and crystallographic analysis of its complex with trypsin.
J.Biochem.(Tokyo), 116, 1994
|
|
4N4G
 
 | |
