2BKT
 
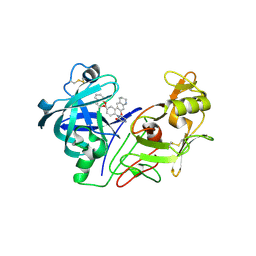 | | crystal structure of renin-pf00257567 complex | | Descriptor: | 1-{4-[3-(2-METHOXY-BENZYLOXY)-PROPOXY]-PHENYL}-6-(1,2,,3,4-TETRAHYDRO-QUINOLIN-7-YLOXYMETHYL)-PIPERAZIN-2-ONE, RENIN | | Authors: | Powell, N.A, Clay, E.H, Holsworth, D.D, Edmunds, J.J, Bryant, J.W, Ryan, J.M, Jalaie, M, Zhang, E. | | Deposit date: | 2005-02-18 | | Release date: | 2006-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Equipotent Activity in Both Enantiomers of a Series of Ketopiperazine-Based Renin Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2I4Q
 
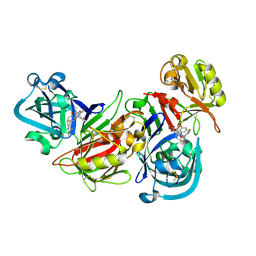 | | Human renin/PF02342674 complex | | Descriptor: | (2S)-6-(2,4-DIAMINO-6-ETHYLPYRIMIDIN-5-YL)-2-(3,5-DIFLUOROPHENYL)-4-(3-METHOXYPROPYL)-2-METHYL-2H-1,4-BENZOXAZIN-3(4H)-ONE, Renin | | Authors: | Holsworth, D.D, Jalaie, M, Zhang, E, Mcconnell, P, Mochalkin, I, Finzel, B.C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design of 6-(2,4-diaminopyrimidinyl)-1,4-benzoxazin-3-ones as small molecule renin inhibitors.
Bioorg.Med.Chem., 15, 2007
|
|
2GRX
 
 | | Crystal structure of TonB in complex with FhuA, E. coli outer membrane receptor for ferrichrome | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-[L-glycero-alpha-D-manno-heptopyranose-(1-5)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2,3-dideoxy-alpha-D-glucoyranose-(1-6)-2-amino-2,3-dideoxy-alpha-D-glucoyranose, ... | | Authors: | Pawelek, P.D, Allaire, M, Coulton, J.W. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of TonB in complex with FhuA, E. coli outer membrane receptor.
Science, 312, 2006
|
|
1PX8
 
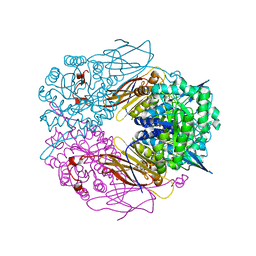 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
3V6E
 
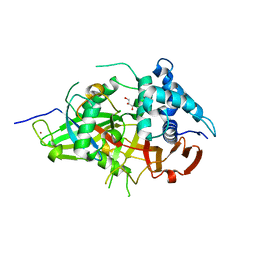 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
2N1U
 
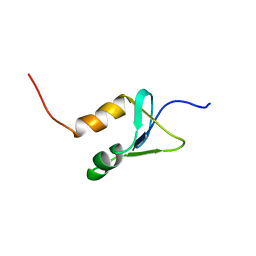 | | Structure of SAP30L corepressor protein | | Descriptor: | Histone deacetylase complex subunit SAP30L, ZINC ION | | Authors: | Tossavainen, H, Permi, P. | | Deposit date: | 2015-04-23 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent disulfide bond formation in SAP30L corepressor protein: Implications for structure and function.
Protein Sci., 25, 2016
|
|
5T2U
 
 | | short chain dehydrogenase/reductase family protein | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Mickael, B, Van Wyk, N, Baneres-Roquet, F, Yann, G, Laurent, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of NADP(+) triggers an open-to-closed transition in a mycobacterial FabG beta-ketoacyl-ACP reductase.
Biochem. J., 474, 2017
|
|
6JRJ
 
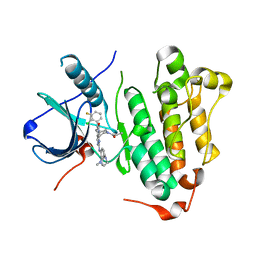 | | The structure of co-crystals of 8r-B-EGFR T790M/C797S complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.943 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
6JRK
 
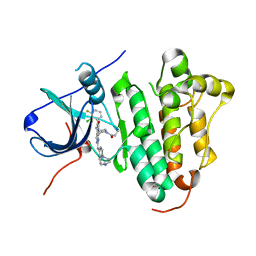 | | The structure of co-crystals of 8r-B-EGFR WT complex | | Descriptor: | 6-(2-chloranyl-3-fluoranyl-phenyl)-5-methyl-2-[[3-methyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-8-[(3S)-1-propanoylpiperidin-3-yl]pyrido[2,3-d]pyrimidin-7-one, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2019-04-04 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure-Based Design of 5-Methylpyrimidopyridone Derivatives as New Wild-Type Sparing Inhibitors of the Epidermal Growth Factor Receptor Triple Mutant (EGFRL858R/T790M/C797S).
J.Med.Chem., 62, 2019
|
|
7VRE
 
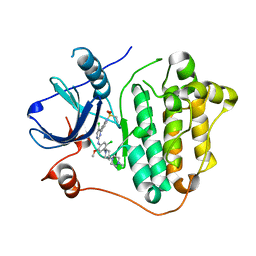 | | The crystal structure of EGFR T790M/C797S with the inhibitor HCD2892 | | Descriptor: | 5-chloranyl-N-[5-chloranyl-2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-4-(1-ethylsulfonylindol-3-yl)pyrimidin-2-amine, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
7VRA
 
 | | The crystal structure of EGFR T790M/C797S with the inhibitor HC5476 | | Descriptor: | 25-chloro-11-(ethylsulfonyl)-44-morpholino-11H-5,12-dioxa-3-aza-1(3,6)-indola-2(4,2)-pyrimidina-4(1,3)-benzenacyclododecaphane, Epidermal growth factor receptor | | Authors: | Zhu, S.J. | | Deposit date: | 2021-10-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Conformational Constrained 4-(1-Sulfonyl-3-indol)yl-2-phenylaminopyrimidine Derivatives as New Fourth-Generation Epidermal Growth Factor Receptor Inhibitors Targeting T790M/C797S Mutations.
J.Med.Chem., 65, 2022
|
|
3FKT
 
 | |
6CCY
 
 | | Crystal structure of Akt1 in complex with a selective inhibitor | | Descriptor: | (5R)-4-(4-{4-[4-fluoro-3-(trifluoromethyl)phenyl]-1-[2-(pyrrolidin-1-yl)ethyl]-1H-imidazol-2-yl}piperidin-1-yl)-5-methyl-5,8-dihydropyrido[2,3-d]pyrimidin-7(6H)-one, RAC-alpha serine/threonine-protein kinase,PIFtide | | Authors: | Wang, Y, Stout, S. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-02 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of chiral dihydropyridopyrimidinones as potent, selective and orally bioavailable inhibitors of AKT.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7ER2
 
 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with LS_2_40 | | Descriptor: | 5-chloranyl-N2-[3-chloranyl-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]-N4-(2-dimethylphosphorylphenyl)pyrimidine-2,4-diamine, Epidermal growth factor receptor | | Authors: | Zhu, S.J, Yun, C.H. | | Deposit date: | 2021-05-05 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Optimization of Brigatinib as New Wild-Type Sparing Inhibitors of EGFR T790M/C797S Mutants.
Acs Med.Chem.Lett., 13, 2022
|
|
5OSG
 
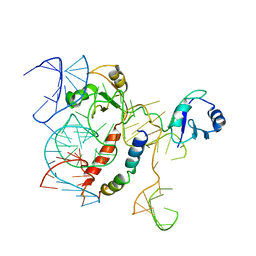 | | Structure of KSRP in context of Leishmania donovani 80S | | Descriptor: | 18S rRNA, 40S ribosomal protein S6, RNA binding protein, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
1S1T
 
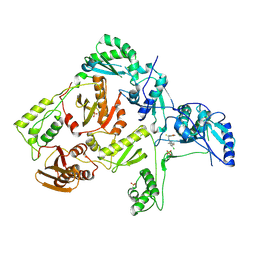 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1X
 
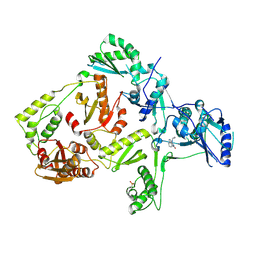 | | Crystal structure of V108I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1V
 
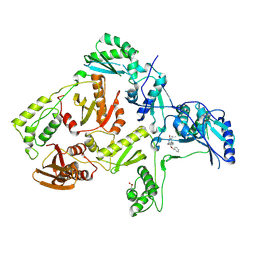 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1W
 
 | | Crystal structure of V106A mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1S1U
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6ENR
 
 | |
7AHA
 
 | | Structure of SARS-CoV-2 Main Protease bound to Maleate. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7AGA
 
 | | Structure of SARS-CoV-2 Main Protease bound to AT7519 | | Descriptor: | 3C-like proteinase, 4-{[(2,6-dichlorophenyl)carbonyl]amino}-N-piperidin-4-yl-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Hinrichs, W, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Boger, J, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
