8GDB
 
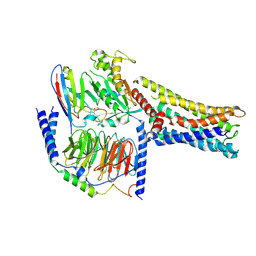 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jianqiang, M, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GDA
 
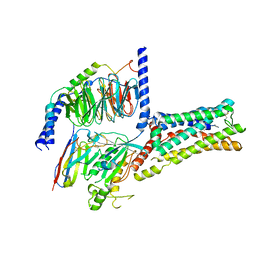 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (2S,5R)-5-[(1E,3S)-4,4-difluoro-3-hydroxy-4-phenylbut-1-en-1-yl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jiangqian, M, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
6WI5
 
 | |
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DMA
 
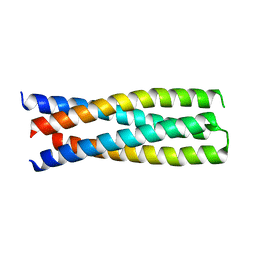 | | DHD15_closed | | Descriptor: | DHD15_closed_A, DHD15_closed_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.363 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DKM
 
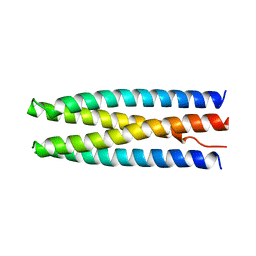 | | DHD131 | | Descriptor: | DHD131_A, DHD131_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DM9
 
 | | DHD15_extended | | Descriptor: | DHD15_extended_A, DHD15_extended_B, SULFATE ION | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DMP
 
 | | De Novo Design of a Protein Heterodimer with Specificity Mediated by Hydrogen Bond Networks | | Descriptor: | Designed orthogonal protein DHD13_XAAA_A, Designed orthogonal protein DHD13_XAAA_B | | Authors: | Chen, Z, Flores-Solis, D, Sgourakis, N.G, Baker, D. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DLC
 
 | |
6MSQ
 
 | | Crystal structure of pRO-2.3 | | Descriptor: | pRO-2.3 | | Authors: | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
6MSR
 
 | | Crystal structure of pRO-2.5 | | Descriptor: | pRO-2.5 | | Authors: | Bick, M.J, Sankaran, B, Boyken, S.E, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
4G2V
 
 | | Structure complex of LGN binding with FRMPD1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, G-protein-signaling modulator 2, ... | | Authors: | Shang, Y, Pan, Z, Wen, W, Wang, W, Zhang, M. | | Deposit date: | 2012-07-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the interaction between LGN and Frmpd1
J.Mol.Biol., 425, 2013
|
|
6M8Q
 
 | | Cleavage and Polyadenylation Specificity Factor Subunit 3 (CPSF3) in complex with NVP-LTM531 | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 3, N-{3,5-dichloro-2-hydroxy-4-[2-(4-methylpiperazin-1-yl)ethoxy]benzene-1-carbonyl}-L-phenylalanine, PHOSPHATE ION, ... | | Authors: | Weihofen, W.A, Salcius, M, Michaud, G. | | Deposit date: | 2018-08-22 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | CPSF3-dependent pre-mRNA processing as a druggable node in AML and Ewing's sarcoma.
Nat.Chem.Biol., 16, 2020
|
|
8XB8
 
 | | The structure of ASFV A137R | | Descriptor: | A137R | | Authors: | Li, C, Song, H, Gao, G.F. | | Deposit date: | 2023-12-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | African swine fever virus A137R assembles into a dodecahedron cage.
J.Virol., 98, 2024
|
|
8Z61
 
 | | Human beta-catenin crystal structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel 1-Phenylpiperidine Urea-Containing Derivatives Inhibiting beta-Catenin/BCL9 Interaction and Exerting Antitumor Efficacy through the Activation of Antigen Presentation of cDC1 Cells.
J.Med.Chem., 67, 2024
|
|
8Z5J
 
 | | Beta-catenin Crystal Structure | | Descriptor: | Catenin beta-1 | | Authors: | Tim, F. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel 1-Phenylpiperidine Urea-Containing Derivatives Inhibiting beta-Catenin/BCL9 Interaction and Exerting Antitumor Efficacy through the Activation of Antigen Presentation of cDC1 Cells.
J.Med.Chem., 67, 2024
|
|
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
6P4I
 
 | | Photoactive Yellow Protein PYP 10ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5F
 
 | | Photoactive Yellow Protein PYP Pure Dark | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5G
 
 | | Photoactive Yellow Protein PYP Dark Full | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
3BLA
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG153249 | | Descriptor: | 5-[(1S)-1-(3-chlorophenyl)ethoxy]quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3BL9
 
 | | Synthetic Gene Encoded DcpS bound to inhibitor DG157493 | | Descriptor: | 5-{[1-(2,3-dichlorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
