7X7P
 
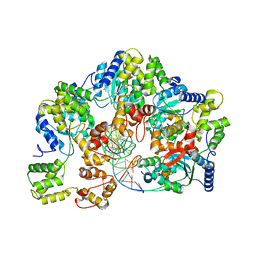 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
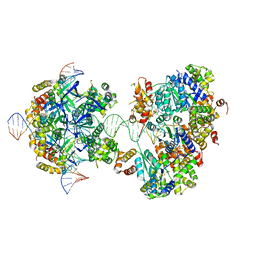 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
8YF1
 
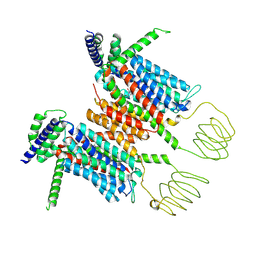 | | Cryo-EM structure of human SV2A in complex with Brivaracetam | | Descriptor: | (2S)-2-[(4R)-2-oxidanylidene-4-propyl-pyrrolidin-1-yl]butanamide, Synaptic vesicle glycoprotein 2A | | Authors: | Qu, Q, Liu, S, Zhou, Z. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Recognition of antiepileptic brivaracetam by synaptic vesicle protein 2A.
Cell Discov, 10, 2024
|
|
8YF0
 
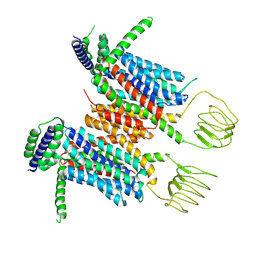 | | Cryo-EM structure of human SV2A | | Descriptor: | Synaptic vesicle glycoprotein 2A | | Authors: | Qu, Q, Liu, S, Zhou, Z. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Recognition of antiepileptic brivaracetam by synaptic vesicle protein 2A.
Cell Discov, 10, 2024
|
|
6KBB
 
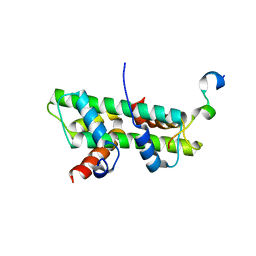 | | Role of the DEF/Y motif of Swc5 in histone H2A.Z deposition | | Descriptor: | Histone H2A type 1-D, Histone H2B type 2-E, SWR1-complex protein 5 | | Authors: | Huang, Y, Zhou, Z. | | Deposit date: | 2019-06-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.365 Å) | | Cite: | Role of a DEF/Y motif in histone H2A-H2B recognition and nucleosome editing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8WLL
 
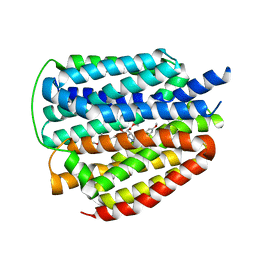 | | Cryo-EM structure of human VMAT2 Y422C, in the presence of reserpine, determined in an inward-facing conformation | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Qu, Q, Wang, Y, Zhou, Z. | | Deposit date: | 2023-09-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Transport and inhibition mechanism for VMAT2-mediated synaptic vesicle loading of monoamines.
Cell Res., 34, 2024
|
|
8WLK
 
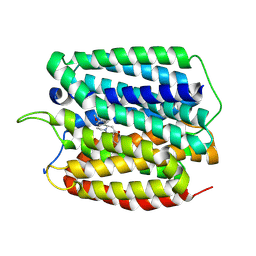 | | Cryo-EM structure of human VMAT2 in presence of Tetrabenazine, determined in an outward-facing conformation | | Descriptor: | (3S,5R,11bS)-9,10-dimethoxy-3-(2-methylpropyl)-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-one, Synaptic vesicular amine transporter | | Authors: | Qu, Q, Wang, Y, Zhou, Z. | | Deposit date: | 2023-09-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Transport and inhibition mechanism for VMAT2-mediated synaptic vesicle loading of monoamines.
Cell Res., 34, 2024
|
|
8WLM
 
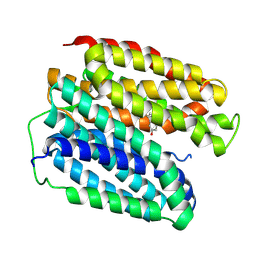 | |
7WLD
 
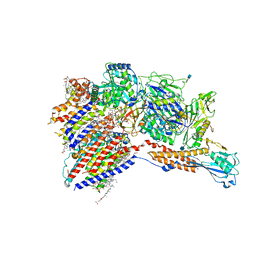 | | Cryo-EM structure of the human glycosylphosphatidylinositol transamidase complex at 2.53 Angstrom resolution | | Descriptor: | (4S,7R)-7-[(hexadecanoyloxy)methyl]-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-3,5,8-trioxa-4lambda~5~-phosphahexacosan-1-aminium, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, Y, Li, T, Luo, Y, Chao, Y, Jia, G, Zhou, Z, Su, Z, Qu, Q, Li, D. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-27 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Molecular insights into biogenesis of glycosylphosphatidylinositol anchor proteins.
Nat Commun, 13, 2022
|
|
7DLX
 
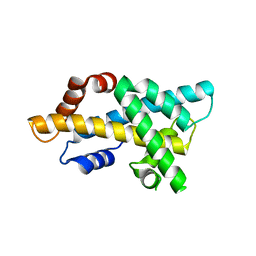 | | crystal structure of H2AM4>Z-H2B | | Descriptor: | Histone H2B,Histone H2A | | Authors: | Dai, L.C, Zhou, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Recognition of the inherently unstable H2A nucleosome by Swc2 is a major determinant for unidirectional H2A.Z exchange.
Cell Rep, 35, 2021
|
|
7XPX
 
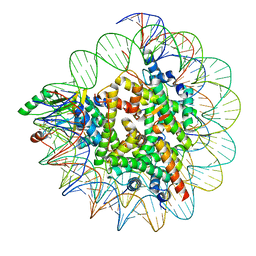 | |
7E8I
 
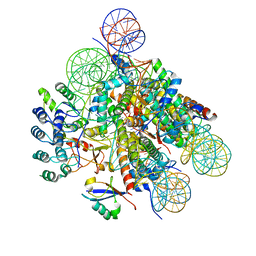 | | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (145-MER), Histone H2A, ... | | Authors: | Dai, Y, Dai, L, Zhou, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin.
Mol.Cell, 81, 2021
|
|
2Q8O
 
 | |
5GT5
 
 | |
7D0E
 
 | | Crystal structure of FIP200 Claw/p-CCPG1 FIR2 | | Descriptor: | 3-(2-hydroxyethyloxy)-2-[2-(2-hydroxyethyloxy)ethoxymethyl]-2-(2-hydroxyethyloxymethyl)propan-1-ol, Cell cycle progression protein 1 FIR2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZG
 
 | | Crystal structure of FIP200 Claw domain apo form | | Descriptor: | DI(HYDROXYETHYL)ETHER, RB1-inducible coiled-coil protein 1 | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-08 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
7CZM
 
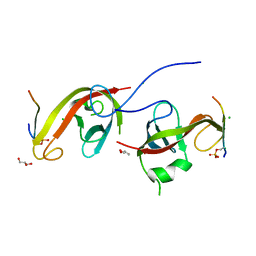 | | Crystal structure of FIP200 Claw/p-OPtineurin LIR complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Optineurin LIR, ... | | Authors: | Zhou, Z.X, Pan, L.F. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation regulates the binding of autophagy receptors to FIP200 Claw domain for selective autophagy initiation.
Nat Commun, 12, 2021
|
|
3JB7
 
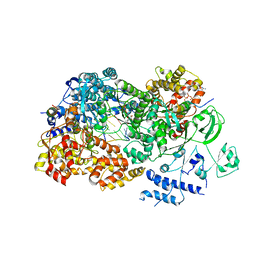 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
6M4G
 
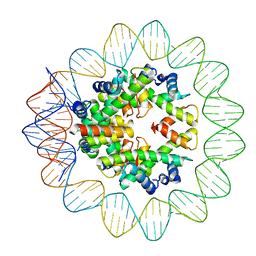 | | Structural mechanism of nucleosome dynamics governed by human histone variants H2A.B and H2A.Z.2.2 | | Descriptor: | DNA (93-MER), Histone H2A-Bbd type 2/3, Histone H2B type 2-E, ... | | Authors: | Zhou, M, Dai, L.C, Li, C.M, Shi, L.X, Huang, Y, Guo, Z.Q. | | Deposit date: | 2020-03-06 | | Release date: | 2020-09-23 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2.
Embo J., 40, 2021
|
|
6M3M
 
 | |
4FZF
 
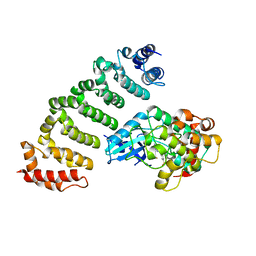 | | Crystal structure of MST4-MO25 complex with DKI | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZA
 
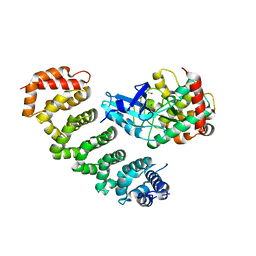 | | Crystal structure of MST4-MO25 complex | | Descriptor: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
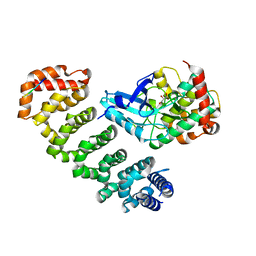 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
6PSN
 
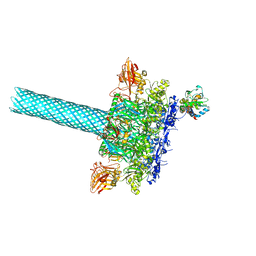 | | Anthrax toxin protective antigen channels bound to lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
4GEH
 
 | |
