4EC5
 
 | |
4J3E
 
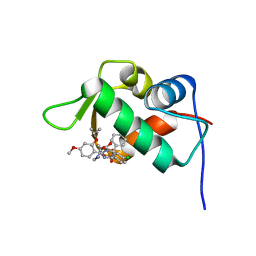 | | The 1.9A crystal structure of humanized Xenopus Mdm2 with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C.M, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development.
ACS Med Chem Lett, 4, 2013
|
|
8XAU
 
 | | Cryo-EM structure of HerA | | Descriptor: | ATP-binding protein | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8XAV
 
 | | Cryo-EM structure of an anti-phage defense complex | | Descriptor: | ATP-binding protein, DUF4297 | | Authors: | Wang, Y, Deng, Z. | | Deposit date: | 2023-12-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex.
Cell Res., 34, 2024
|
|
8II0
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(3-(3-chlorophenyl)isoxazol-5-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[3-(3-chlorophenyl)-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8IHZ
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(1-(3-(4-chlorophenyl)propyl)-1H-1,2,3-triazol-4-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[1-[3-(4-chlorophenyl)propyl]-1,2,3-triazol-4-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8XAX
 
 | |
8XAW
 
 | |
8XAY
 
 | |
8I88
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA complex | | Descriptor: | Piwi domain-containing protein, RNA (5'-R(P*GP*A)-3'), TIR domain-containing protein | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8I87
 
 | | Cryo-EM structure of TIR-APAZ/Ago-gRNA-DNA complex | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*A)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Zhang, H, Deng, Z.Q, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8IN8
 
 | | Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*A)-3'), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7W0S
 
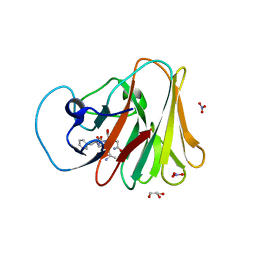 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0Q
 
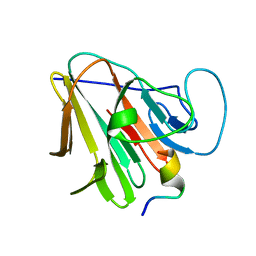 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0T
 
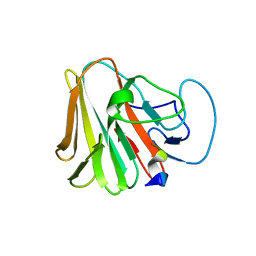 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7, peptide | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7ZRB
 
 | |
7WK0
 
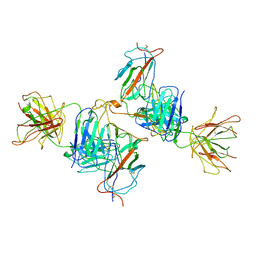 | | Local refine of Omicron spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJY
 
 | | Omicron spike trimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WJZ
 
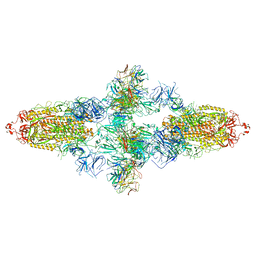 | | Omicron Spike bitrimer with 6m6 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6m6 heavy chain, 6m6 light chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | A broadly neutralizing antibody against SARS-CoV-2 Omicron variant infection exhibiting a novel trimer dimer conformation in spike protein binding.
Cell Res., 32, 2022
|
|
7WOS
 
 | | The state 3 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOQ
 
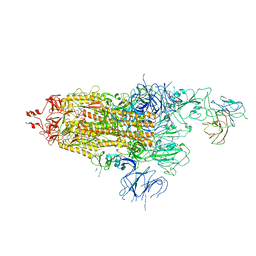 | | The state 1 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOU
 
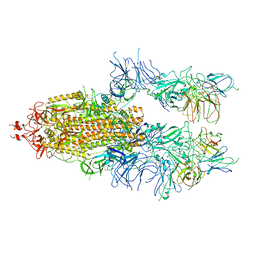 | | The state 4 of Omicron Spike with bispecific antibody FD01 | | Descriptor: | 16L9 Fv, 2-acetamido-2-deoxy-beta-D-glucopyranose, GW01 Fv, ... | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
7WOP
 
 | | The local refined map of Omicron spike with bispecific antibody FD01 | | Descriptor: | 16L9, GW01, Spike protein S1 | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
