4CUG
 
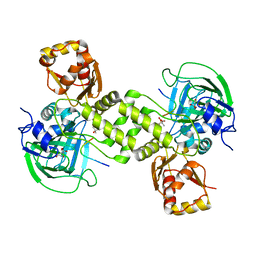 | | Rhodothermus marinus YCFD-like ribosomal protein L16 Arginyl hydroxylase in complex substrate fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S RIBOSOMAL PROTEIN L16, CUPIN 4 FAMILY PROTEIN, ... | | Authors: | McDonough, M.A, Sekirnik, R, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4LIU
 
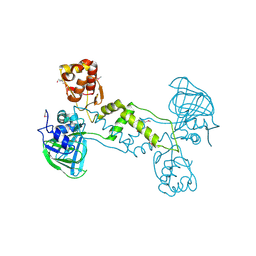 | | Structure of YcfD, a Ribosomal oxygenase from Escherichia coli. | | Descriptor: | 50S ribosomal protein L16 arginine hydroxylase, PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Brissett, N.C, Doherty, A.J, Fox, G.C. | | Deposit date: | 2013-07-03 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 509, 2014
|
|
8ROY
 
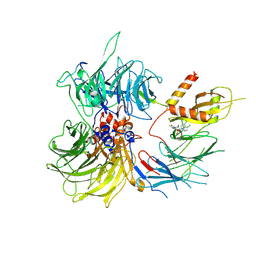 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROX
 
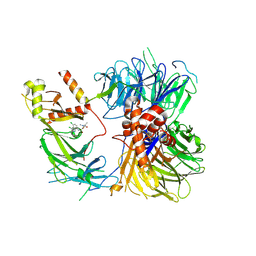 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
4LIV
 
 | |
2V1W
 
 | | Crystal structure of human LIM protein RIL (PDLIM4) PDZ domain bound to the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PDZ AND LIM DOMAIN PROTEIN 4, ... | | Authors: | Soundararajan, M, Shrestha, L, Pike, A.C.W, Salah, E, Burgess-Brown, N, Elkins, J, Umeano, C, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Interactions in Pdz Domain Crystal Structures Help Explain Binding Mechanisms.
Protein Sci., 19, 2010
|
|
2UZC
 
 | | Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ AND LIM DOMAIN 5 | | Authors: | Bunkoczi, G, Elkins, J, Salah, E, Burgess-Brown, N, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms.
Protein Sci., 19, 2010
|
|
9GIO
 
 | |
8OWG
 
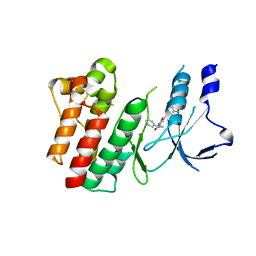 | | Crystal structure of D1228V c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUU
 
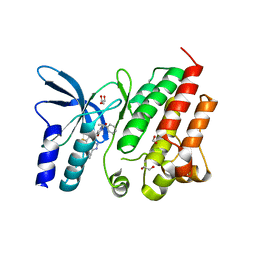 | | Crystal structure of D1228V c-MET bound by compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3-ethynyl-5-fluoranyl-1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, FORMIC ACID, ... | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OVZ
 
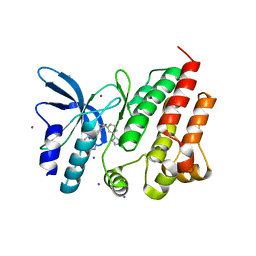 | | Crystal structure of D1228V c-MET bound by compound 16 | | Descriptor: | 1-[(1S)-1-[3-(1H-imidazol-4-yl)phenyl]ethyl]-5-(1H-indazol-7-yl)pyrimidine-2,4-dione, Hepatocyte growth factor receptor, IODIDE ION | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUV
 
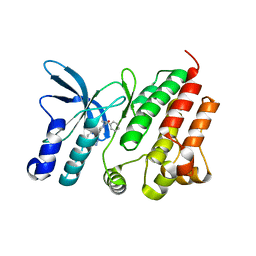 | | Crystal structure of D1228V c-MET bound by compound 15 | | Descriptor: | 5-(1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OW3
 
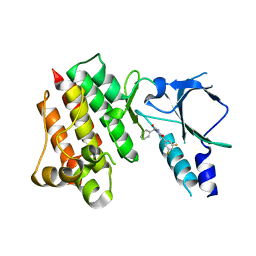 | | Crystal structure of wild-type c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OV7
 
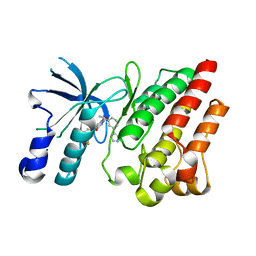 | | Crystal structure of D1228V c-MET bound by compound 10 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-[3-(1H-imidazol-5-yl)phenyl]ethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8G47
 
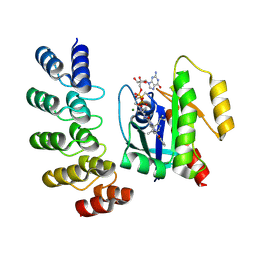 | | KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold | | Descriptor: | AMG 510 (bound form), GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4E
 
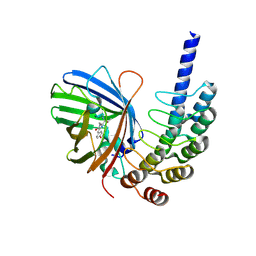 | |
8G3K
 
 | |
8G42
 
 | | KRAS G12C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4H
 
 | | KRAS G13C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4F
 
 | | KRAS G12V complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4DIQ
 
 | | Crystal Structure of human NO66 | | Descriptor: | Lysine-specific demethylase NO66, NICKEL (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Vollmar, M, Krojer, T, Ng, S, Pilka, E, Bray, J, Pike, A.C.W, Filippakopoulos, P, Roos, A, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of human NO66
TO BE PUBLISHED
|
|
4BXF
 
 | |
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCM
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G220C) peptide fragment (complex-1) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CCO
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66 S373C) in complex with Mn(II), N-oxalylglycine (NOG) and 60S ribosomal protein L8 (RPL8 G214C) peptide fragment (complex-3) | | Descriptor: | 1,2-ETHANEDIOL, 60S RIBOSOMAL PROTEIN L8, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66, ... | | Authors: | Chowdhury, R, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
