7CRD
 
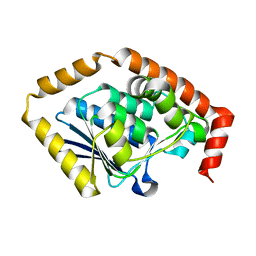 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
4MD6
 
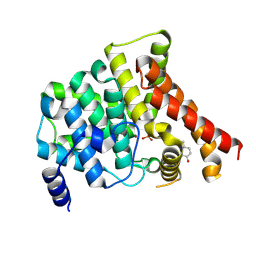 | | Crystal structure of PDE5 in complex with inhibitor 5R | | Descriptor: | 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cui, W, Huang, M, Shao, Y, Luo, H. | | Deposit date: | 2013-08-22 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 3-(4-hydroxybenzyl)-1-(thiophen-2-yl)chromeno[2,3-c]pyrrol-9(2H)-one as a phosphodiesterase-5 inhibitor and its complex crystal structure.
Biochem Pharmacol, 89, 2014
|
|
5ZKQ
 
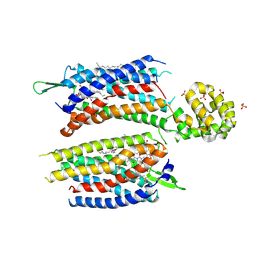 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5ZKP
 
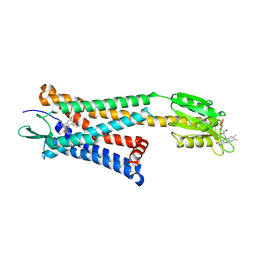 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8JI0
 
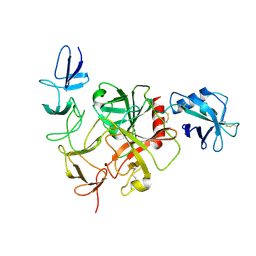 | | Cryo-EM structure of the TcsH-CROP in complex with TMPRSS2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Hemorrhagic toxin, Transmembrane protease serine 2 | | Authors: | Zhou, R, Tao, L, Zhan, X. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of TMPRSS2 recognition by Paeniclostridium sordellii hemorrhagic toxin.
Nat Commun, 15, 2024
|
|
8JXS
 
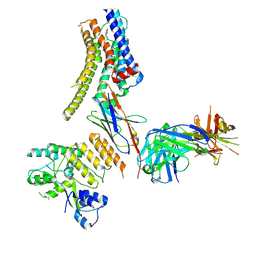 | | Structure of nanobody-bound DRD1_PF-6142 complex | | Descriptor: | 4-[3-methyl-4-(6-methylimidazo[1,2-a]pyrazin-5-yl)phenoxy]furo[3,2-c]pyridine, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 112, 2024
|
|
8JXR
 
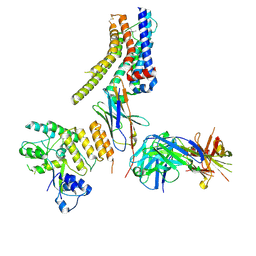 | | Structure of nanobody-bound DRD1_LSD complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 112, 2024
|
|
6K7O
 
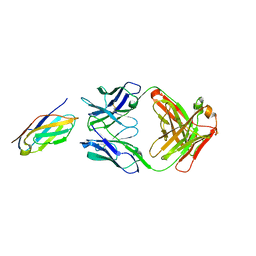 | | Complex structure of LILRB4 and h128-3 antibody | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, h128-3 Fab heavy chain, h128-3 Fab light chain | | Authors: | Song, H, Chai, Y, Xu, X, Gao, F.G. | | Deposit date: | 2019-06-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Disrupting LILRB4/APOE Interaction by an Efficacious Humanized Antibody Reverses T-cell Suppression and Blocks AML Development.
Cancer Immunol Res, 7, 2019
|
|
8JHZ
 
 | | Cryo-EM structure of the TcsH-TMPRSS2 complex | | Descriptor: | Hemorrhagic toxin, Transmembrane protease serine 2, ZINC ION | | Authors: | Zhou, R, Liang, T, Zhan, X. | | Deposit date: | 2023-05-25 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of TMPRSS2 recognition by Paeniclostridium sordellii hemorrhagic toxin.
Nat Commun, 15, 2024
|
|
6D3S
 
 | | Thermostabilized phosphorylated chicken CFTR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator | | Authors: | Fay, J.F, Riordan, J.R, Chen, Z.J. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM Visualization of an Active High Open Probability CFTR Anion Channel.
Biochemistry, 57, 2018
|
|
6D3R
 
 | | Thermostablilized dephosphorylated chicken CFTR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator | | Authors: | Fay, J.F, Riordan, J.R. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM Visualization of an Active High Open Probability CFTR Anion Channel.
Biochemistry, 57, 2018
|
|
7CSW
 
 | | Pseudomonas aeruginosa antitoxin HigA with pa2440 promoter | | Descriptor: | HTH cro/C1-type domain-containing protein, pa2440 | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSV
 
 | | Pseudomonas aeruginosa antitoxin HigA | | Descriptor: | HTH cro/C1-type domain-containing protein | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSY
 
 | | Pseudomonas aeruginosa antitoxin HigA with higBA promoter | | Descriptor: | DNA (28-MER), DNA (29-MER), HTH cro/C1-type domain-containing protein, ... | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
5Y9O
 
 | | The apo structure of Legionella pneumophila WipA | | Descriptor: | IODIDE ION, WipA | | Authors: | Xie, W, Jia, Q. | | Deposit date: | 2017-08-26 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Legionella pneumophila effector WipA, a bacterial PPP protein phosphatase with PTP activity
Acta Biochim. Biophys. Sin. (Shanghai), 50, 2018
|
|
7UMB
 
 | | NanoBRET tracer Tram-bo bound to a KSR2-MEK1 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, Kinase suppressor of Ras 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Marsiglia, W.M, Khan, K.M, Dar, A.C. | | Deposit date: | 2022-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.231 Å) | | Cite: | Live-cell target engagement of allosteric MEKi on MEK-RAF/KSR-14-3-3 complexes.
Nat.Chem.Biol., 20, 2024
|
|
6LOM
 
 | |
8EQN
 
 | |
7JTG
 
 | |
7JTF
 
 | |
7VIG
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIE
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIF
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with (S)-FTY720-P | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VIH
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W8A
 
 | | Babesia orientalis lactate dehydrogenase, BoLDH apo | | Descriptor: | lactate dehydrogenase | | Authors: | Yu, L. | | Deposit date: | 2021-12-07 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | The Structural Basis of Babesia orientalis Lactate Dehydrogenase.
Front Cell Infect Microbiol, 11, 2021
|
|
