6ZN5
 
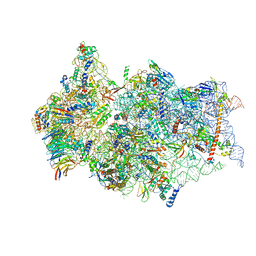 | | SARS-CoV-2 Nsp1 bound to a pre-40S-like ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6ZP4
 
 | | SARS-CoV-2 Nsp1 bound to a human 43S preinitiation ribosome complex - state 2 | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
7ZKY
 
 | |
6YHF
 
 | | Solution NMR Structure of APP TMD | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHO
 
 | | Solution NMR Structure of APP G38P mutant TM | | Descriptor: | Amyloid-beta precursor protein G38P mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHP
 
 | | Solution NMR Structure of APP V44M mutant TMD | | Descriptor: | Amyloid-beta precursor protein V44M mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHX
 
 | | Solution NMR Structure of APP I45T mutant TMD | | Descriptor: | Amyloid-beta precursor protein I45T mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHI
 
 | | Solution NMR Structure of APP G38L mutant TMD | | Descriptor: | Amyloid-beta precursor protein G38L mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
4I5P
 
 | | Selective & Brain-Permeable Polo-like Kinase-2 (Plk-2) Inhibitors that Reduce -Synuclein Phosphorylation in Rat Brain | | Descriptor: | (7R)-8-cyclopentyl-7-ethyl-5-methyl-2-(1H-pyrrol-2-yl)-7,8-dihydropteridin-6(5H)-one, Serine/threonine-protein kinase PLK2 | | Authors: | Aubele, D.L, Hom, R.K, Adler, M, Galemmo Jr, R.A, Bowers, S, Truong, A.P, Pan, H, Beroza, P. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | Selective and brain-permeable polo-like kinase-2 (Plk-2) inhibitors that reduce alpha-synuclein phosphorylation in rat brain.
Chemmedchem, 8, 2013
|
|
4I6H
 
 | |
4I6F
 
 | |
4I6B
 
 | |
6RZV
 
 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (20.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6EL7
 
 | | Glucocorticoid Receptor in complex with compound 31 | | Descriptor: | 1,2-ETHANEDIOL, Glucocorticoid receptor, Nuclear receptor coactivator 2, ... | | Authors: | Edman, K, Wissler, L. | | Deposit date: | 2017-09-28 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of a Novel Oral Glucocorticoid Receptor Modulator (AZD9567) with Improved Side Effect Profile.
J. Med. Chem., 61, 2018
|
|
6RZT
 
 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
8B3X
 
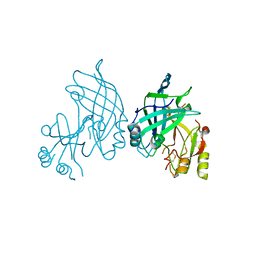 | | High resolution crystal structure of dimeric SUDV VP40 | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Norris, M, Saphire, E.O, Becker, S. | | Deposit date: | 2022-09-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
6RZW
 
 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
5MZK
 
 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-[5-chloro-6-(cyclobutylmethoxy)-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl]propanoic acid | | Descriptor: | 3-[5-chloro-6-(cyclobutylmethoxy)-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl]propanoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Development of a Series of Kynurenine 3-Monooxygenase Inhibitors Leading to a Clinical Candidate for the Treatment of Acute Pancreatitis.
J. Med. Chem., 60, 2017
|
|
5MZC
 
 | |
8B1P
 
 | | Crystal structure of SUDV VP40 CCS mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
8B1O
 
 | | Crystal structure of SUDV VP40 C314S mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
8AW0
 
 | |
8AVZ
 
 | |
5MZI
 
 | | Pseudomonas fluorescens kynurenine 3-monooxygenase (KMO) in complex with 3-(5-chloro-6-cyclopropoxy-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid | | Descriptor: | 3-(5-chloro-6-cyclopropoxy-2-oxo-2,3-dihydro-1,3-benzoxazol-3-yl)propanoic acid, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rowland, P. | | Deposit date: | 2017-01-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Development of a Series of Kynurenine 3-Monooxygenase Inhibitors Leading to a Clinical Candidate for the Treatment of Acute Pancreatitis.
J. Med. Chem., 60, 2017
|
|
5FHV
 
 | | Crystal structure of mCherry after reaction with 2-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, HEXAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2015-12-22 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Efficient switching of mCherry fluorescence using chemical caging.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
