2WJY
 
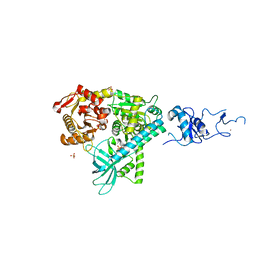 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 Orthorhombic form | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, SULFATE ION, ZINC ION | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
4NAK
 
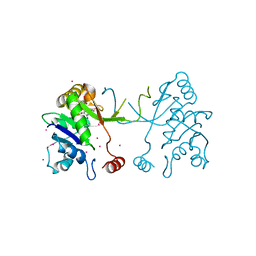 | | Arabidopsis thaliana IspD in complex with pentabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6VXG
 
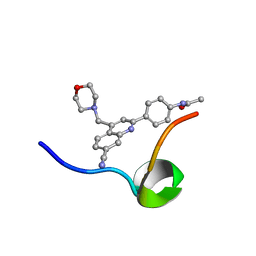 | | Structure of the C-terminal Domain of RAGE and Its Inhibitor | | Descriptor: | Advanced glycosylation end product-specific receptor, N-(4-{7-cyano-4-[(morpholin-4-yl)methyl]quinolin-2-yl}phenyl)acetamide | | Authors: | Ramirez, L, Shekhtman, A. | | Deposit date: | 2020-02-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small-molecule antagonism of the interaction of the RAGE cytoplasmic domain with DIAPH1 reduces diabetic complications in mice.
Sci Transl Med, 13, 2021
|
|
5N2W
 
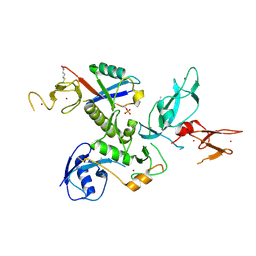 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7LAJ
 
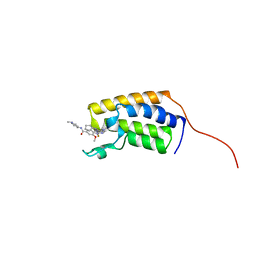 | | Crystal structure of the first bromodomain (BD1) of human BRD2 bound to Ro3280 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 2 | | Authors: | Karim, M.R, Bikowitz, M, Chan, A, Schonbrunn, E. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
4NAN
 
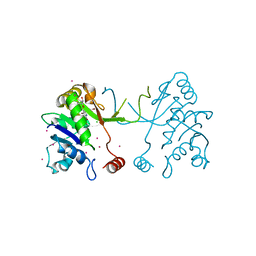 | | Arabidopsis thaliana IspD in complex with tetrabromo-pseudilin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Kunfermann, A, Witschel, M, Illarionov, B, Martin, R, Rottmann, M, Hoffken, H.W, Seet, M, Eisenreich, W, Knolker, H.-J, Fischer, M, Bacher, A, Groll, M, Diederich, F. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pseudilins: Halogenated, Allosteric Inhibitors of the Non-Mevalonate Pathway Enzyme IspD.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2X0L
 
 | | Crystal structure of a neuro-specific splicing variant of human histone lysine demethylase LSD1. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HISTONE H3 PEPTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Zibetti, C, Adamo, A, Binda, C, Forneris, F, Verpelli, C, Ginelli, E, Mattevi, A, Sala, C, Battaglioli, E. | | Deposit date: | 2009-12-15 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative Splicing of the Histone Demethylase Lsd1/Kdm1 Contributes to the Modulation of Neurite Morphogenesis in the Mammalian Nervous System.
J.Neurosci., 30, 2010
|
|
2WVW
 
 | | Cryo-EM structure of the RbcL-RbcX complex | | Descriptor: | RBCX PROTEIN, RIBULOSE BISPHOSPHATE CARBOXYLASE LARGE CHAIN | | Authors: | Liu, C, Young, A.L, Starling-Windhof, A, Bracher, A, Saschenbrecker, S, Rao, B.V, Rao, K.V, Berninghausen, O, Mielke, T, Hartl, F.U, Beckmann, R, Hayer-Hartl, M. | | Deposit date: | 2009-10-20 | | Release date: | 2010-01-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Coupled Chaperone Action in Folding and Assembly of Hexadecameric Rubisco
Nature, 463, 2010
|
|
8AK4
 
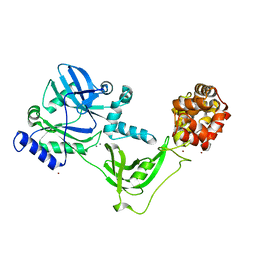 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
6ZDR
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 4d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3NFE
 
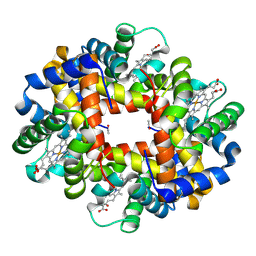 | | The crystal structure of hemoglobin I from trematomus newnesi in deoxygenated state | | Descriptor: | Hemoglobin subunit alpha-1, Hemoglobin subunit beta-1/2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Vitagliano, L, Merlino, A, Sica, F, Marino, K, Mazzarella, L. | | Deposit date: | 2010-06-10 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | An order-disorder transition plays a role in switching off the root effect in fish hemoglobins.
J.Biol.Chem., 285, 2010
|
|
5NFF
 
 | | Crystal structure of GP1 receptor binding domain from Morogoro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Israeli, H, Cohen-Dvashi, H, Shulman, A, Shimon, A, Diskin, R. | | Deposit date: | 2017-03-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Mapping of the Lassa virus LAMP1 binding site reveals unique determinants not shared by other old world arenaviruses.
PLoS Pathog., 13, 2017
|
|
7QNV
 
 | |
7QOB
 
 | |
8B31
 
 | |
5LXF
 
 | | Crystal structure of the human Macrophage Colony Stimulating Factor M- CSF_C31S variant | | Descriptor: | Macrophage colony-stimulating factor 1 | | Authors: | Shahar, A, Papo, N, Zarivach, R, Kosloff, M, Bakhman, A, Rosenfeld, L, Zur, Y, Levaot, N. | | Deposit date: | 2016-09-21 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a monomeric variant of macrophage colony-stimulating factor (M-CSF) that antagonizes the c-FMS receptor.
Biochem. J., 474, 2017
|
|
6FM9
 
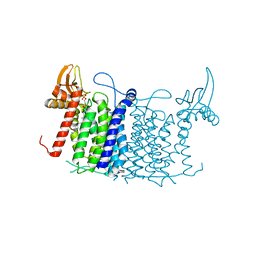 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
7QSI
 
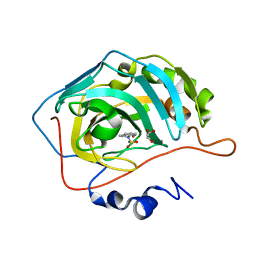 | | human Carbonic Anhydrase II in complex with indoline-1-sulfonamide | | Descriptor: | 2,3-dihydroindole-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-01-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Diversely substituted sulfamides for fragment-based drug discovery of carbonic anhydrase inhibitors: synthesis and inhibitory profile.
J Enzyme Inhib Med Chem, 37, 2022
|
|
7JXT
 
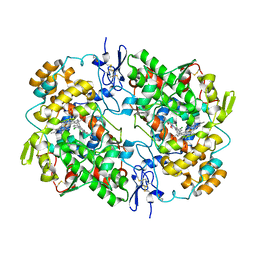 | | Ovine COX-1 in complex with the subtype-selective derivative 2a | | Descriptor: | 2-[4,5-bis(2-chlorophenyl)-1H-imidazol-2-yl]-6-(prop-2-en-1-yl)phenyl methoxyacetate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ko, Y, Iaselli, M, Miciaccia, M, Friedrich, L, Schneider, G, Scilimati, A, Cingolani, G. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Learning from Nature: From a Marine Natural Product to Synthetic Cyclooxygenase-1 Inhibitors by Automated De Novo Design.
Adv Sci, 8, 2021
|
|
5MRC
 
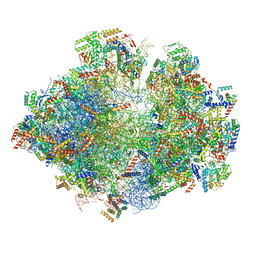 | | Structure of the yeast mitochondrial ribosome - Class A | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2018-02-07 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
3OXP
 
 | | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution | | Descriptor: | GLYCEROL, Phosphotransferase enzyme II, A component, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-21 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of phosphotransferase enzyme II, A component from Yersinia pestis CO92 at 1.2 A resolution
To be Published
|
|
2NR6
 
 | | Crystal structure of the complex of antibody and the allergen Bla g 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody heavy chain, ... | | Authors: | Li, M, Gustchina, A, Wlodawer, A, Pomes, A, Wunschmann, S. | | Deposit date: | 2006-11-01 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody.
J.Biol.Chem., 283, 2008
|
|
4ND9
 
 | | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation | | Descriptor: | ABC transporter, substrate binding protein (Proline/glycine/betaine) | | Authors: | Nicholls, R, Tkaczuk, K.L, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation
To be Published
|
|
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7QZ1
 
 | | Formate dehydrogenase from Starkeya novella | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Pontillo, N, Slotboom, D.J, Guskov, A. | | Deposit date: | 2022-01-30 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural insight into the chemical resistance and cofactor specificity of the formate dehydrogenase from Starkeya novella.
Febs J., 290, 2023
|
|
