4WJ4
 
 | | Crystal structure of non-discriminating aspartyl-tRNA synthetase from Pseudomonas aeruginosa complexed with tRNA(Asn) and aspartic acid | | Descriptor: | 76mer-tRNA, ASPARTIC ACID, Aspartate--tRNA(Asp/Asn) ligase | | Authors: | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2RKG
 
 | | HIV-1 PR resistant mutant + LPV | | Descriptor: | GLYCEROL, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, PROTEASE RETROPEPSIN | | Authors: | Rezacova, P, Brynda, J, Kozisek, M, Saskova, K, Konvalinka, J. | | Deposit date: | 2007-10-16 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ninety-nine is not enough: molecular characterization of inhibitor-resistant human immunodeficiency virus type 1 protease mutants with insertions in the flap region
J.Virol., 82, 2008
|
|
7Z3B
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, reduced form | | Descriptor: | ACETATE ION, AcoP, COPPER (I) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
6Q6F
 
 | | Crystal structure of IDH1 R132H in complex with HMS101 | | Descriptor: | (2~{R})-2-[2-[(3~{R})-3-(4-fluorophenyl)pyrrolidin-1-yl]ethyl]-1,4-dimethyl-piperazine, Isocitrate dehydrogenase [NADP] cytoplasmic | | Authors: | Chaturvedi, A, Goparaju, R, Gupta, C, Kluenemann, T, Araujo Cruz, M.M, Kloos, A, Goerlich, K, Schottmann, R, Struys, E.A, Ganser, A, Preller, M, Heuser, M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site.
Leukemia, 34, 2020
|
|
2SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water molecules participate in proteinase-inhibitor interactions: crystal structures of Leu18, Ala18, and Gly18 variants of turkey ovomucoid inhibitor third domain complexed with Streptomyces griseus proteinase B.
Protein Sci., 4, 1995
|
|
7Z3F
 
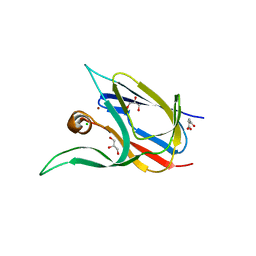 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, oxidized form | | Descriptor: | ACETATE ION, AcoP, CHLORIDE ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3G
 
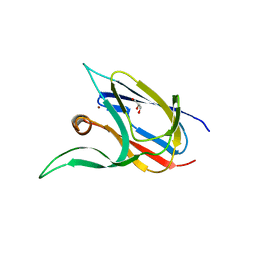 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, H166A mutant | | Descriptor: | AcoP, COPPER (I) ION, GLYCEROL | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
1SI8
 
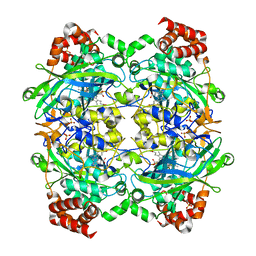 | | Crystal structure of E. faecalis catalase | | Descriptor: | CHLORIDE ION, Catalase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hakansson, K.O, Brugna, M, Tasse, L. | | Deposit date: | 2004-02-28 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of catalase from Enterococcus faecalis.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7Z3I
 
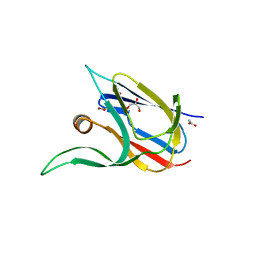 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, M171A mutant | | Descriptor: | ACETATE ION, AcoP, COPPER (II) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
4WSZ
 
 | |
4WT0
 
 | |
7B2L
 
 | | Structure of the endocytic adaptor complex AENTH | | Descriptor: | ANTH domain of Sla2, ENTH domain of epsin Ent1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Klebl, D.P, Lizarrondo, J, Sobott, F, Garcia-Alai, M, Muench, S.P. | | Deposit date: | 2020-11-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the endocytic adaptor complex reveals the basis for efficient membrane anchoring during clathrin-mediated endocytosis.
Nat Commun, 12, 2021
|
|
6QAA
 
 | | Human Butyrylcholinesterase in complex with (S)-2-(butylamino)-N-(2-cycloheptylethyl)-3-(1H-indol-3-yl)propanamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Nachon, F, Harst, M, Knez, D, Gobec, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Tryptophan-derived butyrylcholinesterase inhibitors as promising leads against Alzheimer's disease.
Chem.Commun.(Camb.), 55, 2019
|
|
4WT3
 
 | | The N-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT4
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form I | | Descriptor: | PHOSPHATE ION, Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WT5
 
 | | The C-terminal domain of Rubisco Accumulation Factor 1 from Arabidopsis thaliana, crystal form II | | Descriptor: | Rubisco Accumulation Factor 1, isoform 2 | | Authors: | Hauser, T, Bhat, J.Y, Milicic, G, Wendler, P, Hartl, F.U, Bracher, A, Hayer-Hartl, M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structure and mechanism of the Rubisco-assembly chaperone Raf1.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3M7Q
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean sea anemone stichodactyla helianthus in complex with bovine pancreatic trypsin | | Descriptor: | Cationic trypsin, Kunitz-type proteinase inhibitor SHPI-1, PHOSPHATE ION | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Gil, D, Talavera, A, Gonzalez, Y, de los angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-03-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
J.Struct.Biol., 180, 2012
|
|
4WM9
 
 | | Acinetobacter baumanii OXA-24 complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Mangani, S, Benvenuti, M, Docquier, J.-D. | | Deposit date: | 2014-10-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis of Selective Inhibition and Slow Reversibility of Avibactam against Class D Carbapenemases: A Structure-Guided Study of OXA-24 and OXA-48.
Acs Chem.Biol., 10, 2015
|
|
6QAE
 
 | | Human Butyrylcholinesterase in complex with (S)-N2-butyl-N1-(2-cycloheptylethyl)-3-(1H-indol-3-yl)-N1,N2-dimethylpropane-1,2-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Nachon, F, Harst, M, Knez, D, Gobec, S. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Tryptophan-derived butyrylcholinesterase inhibitors as promising leads against Alzheimer's disease.
Chem.Commun.(Camb.), 55, 2019
|
|
4WNJ
 
 | | Crystal structure of Transthyretin-quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Berni, R, Folli, C. | | Deposit date: | 2014-10-13 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1INH
 
 | | INFLUENZA A SUBTYPE N2 NEURAMINIDASE COMPLEXED WITH AROMATIC BANA111 INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(ACETYLAMINO)-3-[(AMINOACETYL)AMINO]BENZOIC ACID, CALCIUM ION, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1995-07-07 | | Release date: | 1996-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitors of influenza virus sialidase. A benzoic acid lead with novel interaction.
J.Med.Chem., 38, 1995
|
|
5HTD
 
 | | Recombinant bovine beta-lactoglobulin variant L1A/I2S with endogenous ligand (sBlgB#1) | | Descriptor: | Beta-lactoglobulin, MYRISTIC ACID | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-01-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
1IRJ
 
 | | Crystal Structure of the MRP14 complexed with CHAPS | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, Migration Inhibitory Factor-Related Protein 14 | | Authors: | Itou, H, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2001-10-09 | | Release date: | 2002-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of human MRP14 (S100A9), a Ca(2+)-dependent regulator protein in inflammatory process.
J.Mol.Biol., 316, 2002
|
|
1ISJ
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with NMN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
4X71
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A269T | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Kanteev, M, Dror, A, Gihaz, S, Shahar, A, Fishman, A. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into methanol-stable variants of lipase T6 from Geobacillus stearothermophilus.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
