5ZEV
 
 | |
5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2025-02-12 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
2MJV
 
 | |
7MCH
 
 | |
7MC4
 
 | |
8ZVF
 
 | | AtALMT9 plus high malate in low pH | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, Aluminum-activated malate transporter 9 | | Authors: | Gong, D.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural insight into the Arabidopsis vacuolar anion channel ALMT9 shows clade specificity.
Cell Rep, 43, 2024
|
|
4NA9
 
 | | Factor VIIa in complex with the inhibitor 3'-amino-5'-[(2s,4r)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid | | Descriptor: | 3'-amino-5'-[(2S,4R)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid, CALCIUM ION, Coagulation factor VII heavy chain, ... | | Authors: | Wei, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors.
J.Med.Chem., 57, 2014
|
|
5J8J
 
 | | A histone deacetylase from Saccharomyces cerevisiae | | Descriptor: | Histone deacetylase HDA1 | | Authors: | Zhu, Y, Shen, H, Li, X, Teng, M. | | Deposit date: | 2016-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.716 Å) | | Cite: | Structural and histone binding ability characterization of the ARB2 domain of a histone deacetylase Hda1 from Saccharomyces cerevisiae.
Sci Rep, 6, 2016
|
|
6AAS
 
 | | Solution Structure for helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (28-MER) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
3OF6
 
 | | Human pre-T cell receptor crystal structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pre T-cell antigen receptor alpha, T cell receptor beta chain | | Authors: | Pang, S.S. | | Deposit date: | 2010-08-13 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Nature, 467, 2010
|
|
2N51
 
 | |
5UCA
 
 | |
8J3Y
 
 | | Crystal structure of CBM6E E168Q in complex with oligosaccharides | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative polysaccharide-binding protein, ... | | Authors: | He, C, Li, F. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into curdlan degradation via a glycoside hydrolase containing a disruptive carbohydrate-binding module.
Biotechnol Biofuels Bioprod, 17, 2024
|
|
8J3X
 
 | | Crystal structure of CBM6E from Saccharophagus degradans | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative polysaccharide-binding protein | | Authors: | He, C, Li, F. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into curdlan degradation via a glycoside hydrolase containing a disruptive carbohydrate-binding module.
Biotechnol Biofuels Bioprod, 17, 2024
|
|
4ETK
 
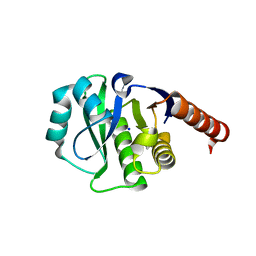 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
6AIB
 
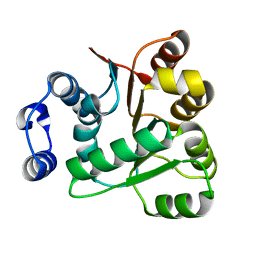 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6AIC
 
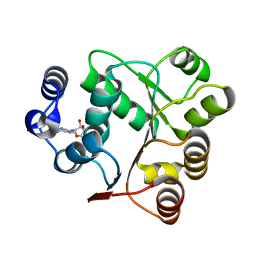 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | Authors: | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5WQJ
 
 | |
6W7F
 
 | | Structure of EED bound to inhibitor 5285 | | Descriptor: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6W7G
 
 | | Structure of EED bound to inhibitor 1056 | | Descriptor: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
5WQK
 
 | |
7EK4
 
 | | prawn ferritin to coordinate with heavy metal ions | | Descriptor: | FE (III) ION, Ferritin, MERCURY (II) ION | | Authors: | Wang, Y, Zang, J. | | Deposit date: | 2021-04-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights for the Stronger Ability of Shrimp Ferritin to Coordinate with Heavy Metal Ions as Compared to Human H-Chain Ferritin.
Int J Mol Sci, 22, 2021
|
|
7EK7
 
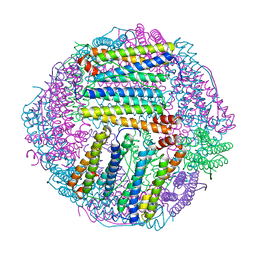 | | prawn ferritin to coordinate with heavy metal ions | | Descriptor: | Ferritin, MERCURY (II) ION | | Authors: | Wang, Y, Zang, J. | | Deposit date: | 2021-04-04 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights for the Stronger Ability of Shrimp Ferritin to Coordinate with Heavy Metal Ions as Compared to Human H-Chain Ferritin.
Int J Mol Sci, 22, 2021
|
|
7EK5
 
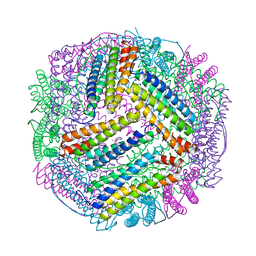 | | prawn ferritin to coordinate with heavy metal ions | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin | | Authors: | Wang, Y, Zang, J. | | Deposit date: | 2021-04-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights for the Stronger Ability of Shrimp Ferritin to Coordinate with Heavy Metal Ions as Compared to Human H-Chain Ferritin.
Int J Mol Sci, 22, 2021
|
|
