1DV4
 
 | | PARTIAL STRUCTURE OF 16S RNA OF THE SMALL RIBOSOMAL SUBUNIT FROM THERMUS THERMOPHILUS | | Descriptor: | 16S RIBOSOMAL RNA, OCTADECATUNGSTENYL DIPHOSPHATE, RIBOSOMAL PROTEIN S5, ... | | Authors: | Tocilj, A, Schlunzen, F, Janell, D, Gluhmann, M, Hansen, H, Harms, J, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The small ribosomal subunit from Thermus thermophilus at 4.5 A resolution: pattern fittings and the identification of a functional site.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3L6R
 
 | | The structure of mammalian serine racemase: Evidence for conformational changes upon inhibitor binding | | Descriptor: | MALONATE ION, MANGANESE (II) ION, Serine racemase | | Authors: | Smith, M.A, Mack, V, Ebneth, A, Cesura, A, Felicetti, B, Barker, J. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-26 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
2FE7
 
 | | The crystal structure of a probable N-acetyltransferase from Pseudomonas aeruginosa | | Descriptor: | probable N-acetyltransferase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a N-acetyltransferase from Pseudomonas aeruginosa
To be Published
|
|
3LAC
 
 | | Crystal structure of Bacillus anthracis pyrrolidone-carboxylate peptidase, pcP | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Pyrrolidone-carboxylate peptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-06 | | Release date: | 2010-01-19 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus anthracis pyrrolidone-carboxylate peptidase, pcP
To be Published
|
|
2FIW
 
 | | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris | | Descriptor: | ACETYL COENZYME *A, GCN5-related N-acetyltransferase:Aminotransferase, class-II, ... | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-30 | | Release date: | 2006-02-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the GCN5-Related N-acetyltransferase: Aminotransferase, Class-II from Rhodopseudomonas palustris
To be Published, 2006
|
|
3LMW
 
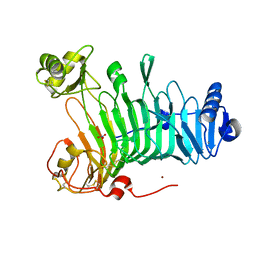 | | Crystal structure of iota-carrageenase family GH82 from A. fortis in absence of chloride ions | | Descriptor: | CALCIUM ION, Iota-carrageenase, CgiA, ... | | Authors: | Rebuffet, E, Barbeyron, T, Jeudy, A, Czjzek, M, Michel, G. | | Deposit date: | 2010-02-01 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of catalytic residues and mechanistic analysis of family GH82 iota-carrageenases
Biochemistry, 49, 2010
|
|
1ZJH
 
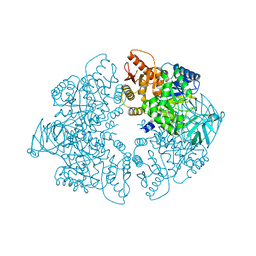 | | Structure of human muscle pyruvate kinase (PKM2) | | Descriptor: | Pyruvate kinase, isozymes M1/M2 | | Authors: | Choe, J, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-04-28 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human muscle pyruvate kinase (PKM2).
TO BE PUBLISHED
|
|
3LPU
 
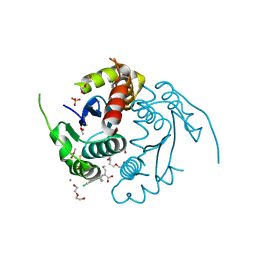 | | HIV integrase | | Descriptor: | (2S)-2-(6-chloro-2-methyl-4-phenylquinolin-3-yl)pentanoic acid, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, CALCIUM ION, ... | | Authors: | Nicolet, S, Christ, F, Voet, A, Marchand, A, Strelkov, S.V, de Maeyer, M, Chaltin, P, Debyzer, Z. | | Deposit date: | 2010-02-06 | | Release date: | 2010-05-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication.
Nat.Chem.Biol., 6, 2010
|
|
1WER
 
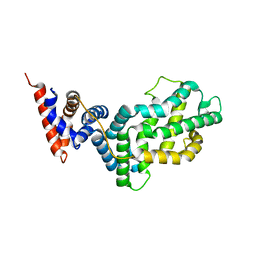 | | RAS-GTPASE-ACTIVATING DOMAIN OF HUMAN P120GAP | | Descriptor: | P120GAP | | Authors: | Scheffzek, K, Lautwein, A, Kabsch, W, Ahmadian, M.R, Wittinghofer, A. | | Deposit date: | 1996-11-20 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras.
Nature, 384, 1996
|
|
2WAO
 
 | | Structure of a family two carbohydrate esterase from Clostridium thermocellum in complex with cellohexaose | | Descriptor: | ENDOGLUCANASE E, FORMIC ACID, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-10 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
3LR2
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Johansson, J, Knight, S.D, Rising, A, Casals, C, Saenz, A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LSO
 
 | |
3LU2
 
 | | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase | | Descriptor: | Lmo2462 protein, ZINC ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
To be Published
|
|
3L6B
 
 | | X-ray crystal structure of human serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Barker, J, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Cesura, A. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
3L6Q
 
 | | Crystal structure of the N-terminal domain of HSP70 from Cryptosporidium parvum (CGD2_20) | | Descriptor: | Heat shock 70 (HSP70) protein, MAGNESIUM ION, SULFATE ION | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Lew, J, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of the N-terminal domain of HSP70 from Cryptosporidium parvum (CGD2_20)
To be Published
|
|
2QQ2
 
 | | Crystal structure of C-terminal domain of Human acyl-CoA thioesterase 7 | | Descriptor: | Cytosolic acyl coenzyme A thioester hydrolase | | Authors: | Busam, R, Lehtio, L, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Herman, M.D, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-26 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human acyl-CoA thioesterase 7.
To be Published
|
|
4AHQ
 
 | | Crystal Structure of N-acetylneuraminic acid lyase mutant K165C from Staphylococcus aureus | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Polyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Trinh, A.R, Berry, A. | | Deposit date: | 2012-02-06 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
2FDR
 
 | | Crystal Structure of Conserved Haloacid Dehalogenase-like Protein of Unknown Function ATU0790 from Agrobacterium tumefaciens str. C58 | | Descriptor: | MAGNESIUM ION, conserved hypothetical protein | | Authors: | Nocek, B, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hydrolases/phosphatases-like fold protein
from Agrobacterium tumefaciens str. C58
To be Published
|
|
3LIT
 
 | | The crystal structure of htlv protease complexed with the inhibitor KNI-10681 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-[(2R)-3-methylbutan-2-yl]-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LPT
 
 | | HIV integrase | | Descriptor: | (6-chloro-2-oxo-4-phenyl-1,2-dihydroquinolin-3-yl)acetic acid, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, CALCIUM ION, ... | | Authors: | Nicolet, S, Christ, F, Voet, A, Marchand, A, Strelkov, S.V, de Maeyer, M, Chaltin, P, Debyzer, Z. | | Deposit date: | 2010-02-05 | | Release date: | 2010-05-12 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication.
Nat.Chem.Biol., 6, 2010
|
|
3LR6
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | Major ampullate spidroin 1, TRIETHYLENE GLYCOL | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Saenz, A, Casals, C, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LUQ
 
 | | The Crystal Structure of a PAS Domain from a Sensory Box Histidine Kinase Regulator from Geobacter sulfurreducens to 2.5A | | Descriptor: | SULFATE ION, Sensor protein, TRIETHYLENE GLYCOL | | Authors: | Stein, A.J, Weger, A, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The Crystal Structure of a PAS Domain from a Sensory Box Histidine Kinase Regulator from Geobacter sulfurreducens to 2.5A
To be Published
|
|
3LVK
 
 | | Crystal Structure of E.coli IscS-TusA complex (form 2) | | Descriptor: | Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE, Sulfurtransferase tusA | | Authors: | Shi, R, Proteau, A, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions.
Plos Biol., 8, 2010
|
|
2XPC
 
 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | Descriptor: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
3LRH
 
 | |
