3S4Y
 
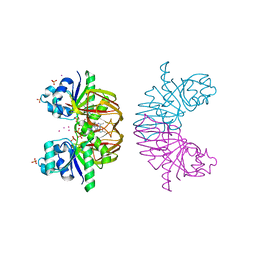 | | Crystal structure of human thiamin pyrophosphokinase 1 | | Descriptor: | CALCIUM ION, SULFATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Shen, L, Tempel, W, Tong, Y, Li, Y, Walker, J.R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human thiamin pyrophosphokinase 1
to be published
|
|
5CQ5
 
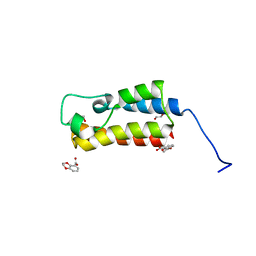 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 2,3-Ethylenedioxybenzoic Acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 2,3-Ethylenedioxybenzoic Acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
4CSH
 
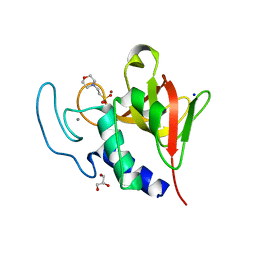 | | Native structure of the lytic CHAPK domain of the endolysin LysK from Staphylococcus aureus bacteriophage K | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Sanz-Gaitero, M, Keary, R, Garcia-Doval, C, Coffey, A, van Raaij, M.J. | | Deposit date: | 2014-03-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the lytic CHAP(K) domain of the endolysin LysK from Staphylococcus aureus bacteriophage K.
Virol. J., 11, 2014
|
|
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3SI1
 
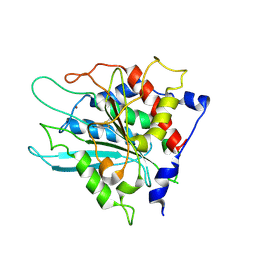 | | Structure of glycosylated murine glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Dambe, T, Carrillo, D, Parthier, C, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
5CYI
 
 | | CDK2/Cyclin A covalent complex with 6-(cyclohexylmethoxy)-N-(4-(vinylsulfonyl)phenyl)-9H-purin-2-amine (NU6300) | | Descriptor: | 6-(cyclohexylmethoxy)-N-[4-(ethylsulfonyl)phenyl]-9H-purin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Anscombe, E, Meschini, E, Vidal, R.M, Martin, M.P, Staunton, D, Geitmann, M, Danielson, U.H, Stanley, W.A, Wang, L.Z, Reuillon, T, Golding, B.T, Cano, C, Newell, D.R, Noble, M.E.M, Wedge, S.R, Endicott, J.A, Griffin, R.J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Characterization of an Irreversible Inhibitor of CDK2.
Chem.Biol., 22, 2015
|
|
5D1B
 
 | | Crystal structure of G117E HDAC8 in complex with TSA | | Descriptor: | Histone deacetylase 8, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Decroos, C, Christianson, N.H, Gullett, L.E, Bowman, C.M, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical and Structural Characterization of HDAC8 Mutants Associated with Cornelia de Lange Syndrome Spectrum Disorders.
Biochemistry, 54, 2015
|
|
5D1D
 
 | | Crystal structure of P91L-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | HDAC8 Fluor de Lys tetrapeptide substrate, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Christianson, N.H, Gullett, L.E, Bowman, C.M, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Biochemical and Structural Characterization of HDAC8 Mutants Associated with Cornelia de Lange Syndrome Spectrum Disorders.
Biochemistry, 54, 2015
|
|
7RZC
 
 | | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor | | Descriptor: | (1R)-N-[(1H-indol-3-yl)methyl]-N-methyl-1-(naphthalen-1-yl)ethan-1-amine, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Wang, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with Jun9-84-3 inhibitor
To be Published
|
|
5CQ4
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 3'-Hydroxyacetophenone (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-hydroxyphenyl)ethanone, Bromodomain adjacent to zinc finger domain protein 2B, ... | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 3'-Hydroxyacetophenone (SGC - Diamond I04-1 fragment screening)
To be published
|
|
5D2B
 
 | | Crystal structure of a mutated catalytic domain of Human MMP12 in complex with an hydroxamate analogue of RXP470 | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-[(2R)-2-{[3-(3'-chlorobiphenyl-4-yl)-1,2-oxazol-5-yl]methyl}-4-(hydroxyamino)-4-oxobutanoyl]-L-alpha-glutamyl-L-alpha-glutamine, ... | | Authors: | Rouanet-Mehouas, C, Devel, L, Dive, V, Stura, E.A. | | Deposit date: | 2015-08-05 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Zinc-Metalloproteinase Inhibitors: Evaluation of the Complex Role Played by the Zinc-Binding Group on Potency and Selectivity.
J. Med. Chem., 60, 2017
|
|
7RTY
 
 | |
3S95
 
 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
3SBL
 
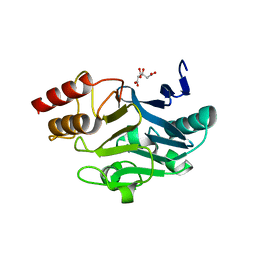 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3SC7
 
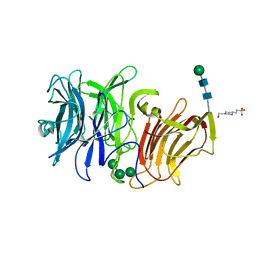 | | First crystal structure of an endo-inulinase, from Aspergillus ficuum: structural analysis and comparison with other GH32 enzymes. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Inulinase, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, ... | | Authors: | Housen, I, Pouyez, J, Roussel, G, Mayard, A, Vandamme, A.M, Wouters, J, Michaux, C. | | Deposit date: | 2011-06-07 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | First crystal structure of an endo-inulinase, INU2, from Aspergillus ficuum: Discovery of an extra-pocket in the catalytic domain responsible for its endo-activity.
Biochimie, 94, 2012
|
|
5DDJ
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus O1M-S2093Y empty capsid | | Descriptor: | Foot and mouth disease virus, VP1, VP2, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7S6O
 
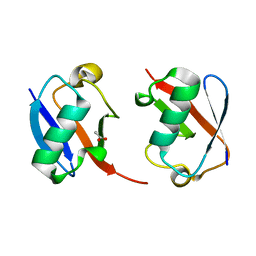 | | The crystal structure of Lys48-linked di-ubiquitin | | Descriptor: | ACETATE ION, Ubiquitin | | Authors: | Osipiuk, J, Tesar, C, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
5DDZ
 
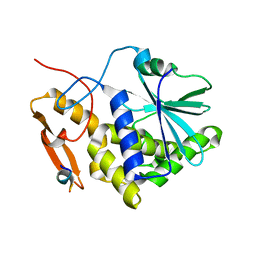 | | Crystal structure of the RTA-c10-P2 complex | | Descriptor: | 60S acidic ribosomal protein P2, Ricin | | Authors: | Zhu, Y, Fan, X, Wang, C, Niu, L, Li, X, Teng, M. | | Deposit date: | 2015-08-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the interaction of the ribosomal P stalk protein P2 with a type II ribosome-inactivating protein ricin
Sci Rep, 6, 2016
|
|
7S6P
 
 | | The crystal structure of human ISG15 | | Descriptor: | Ubiquitin-like protein ISG15 | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
5D8A
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093F empty capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-16 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3SN5
 
 | | Crystal structure of human CYP7A1 in complex with cholest-4-en-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, Cholesterol 7-alpha-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Strushkevich, N, Tempel, W, MacKenzie, F, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of human CYP7A1 in complex with cholest-4-en-3-one
To be Published
|
|
5CRG
 
 | |
3SI0
 
 | | Structure of glycosylated human glutaminyl cyclase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D, Stubbs, M.T. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Glycosylated Mammalian Glutaminyl Cyclases Reveal Conformational Variability near the Active Center.
Biochemistry, 50, 2011
|
|
5CXL
 
 | |
5CTP
 
 | | Crystal structure of CK2alpha with N-(3-(3-chloro-4-(phenyl)benzylamino)propyl)acetamide bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, N-(3-{[(2-chlorobiphenyl-4-yl)methyl]amino}propyl)acetamide, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
