5UFR
 
 | | Structure of RORgt bound to | | Descriptor: | (S)-[4-chloro-2-(dimethylamino)-3-phenylquinolin-6-yl](1-methyl-1H-imidazol-5-yl)(pyridin-4-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6CVH
 
 | | Identification and biological evaluation of thiazole-based inverse agonists of RORgt | | Descriptor: | Nuclear receptor ROR-gamma, trans-3-({4-(cyclohexylmethyl)-5-[3-(1-methylcyclopropyl)-5-{[(2R)-1,1,1-trifluoropropan-2-yl]carbamoyl}phenyl]-1,3-thiazole-2-carbonyl}amino)cyclobutane-1-carboxylic acid | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification and biological evaluation of thiazole-based inverse agonists of ROR gamma t.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6VSW
 
 | | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of RORgt | | Descriptor: | 5-(2,3-dichloro-4-{[(2S)-1,1,1-trifluoropropan-2-yl]sulfamoyl}phenyl)-4-(4-fluoropiperidine-1-carbonyl)-N-(2-hydroxy-2-methylpropyl)-1,3-thiazole-2-carboxamide, RAR-related orphan receptor C | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2020-02-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of ROR gamma t.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
1T4V
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | Hirudin IIIA, N-ALLYL-5-AMIDINOAMINOOXY-PROPYLOXY-3-CHLORO-N-CYCLOPENTYLBENZAMIDE, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
1T4U
 
 | | Crystal Structure Analysis of a novel Oxyguanidine bound to Thrombin | | Descriptor: | 2-METHANESULFONYL-BENZENESULFONIC ACID 3-METHYL-5-((1-AMIDINOAMINOOXYMETHYL-CYCLOPROPYL)METHYLOXY)-PHENYLESTER, Hirudin IIIA, Prothrombin | | Authors: | Spurlino, J. | | Deposit date: | 2004-04-30 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxyguanidines. Part 2: Discovery of a novel orally active thrombin inhibitor through structure-based drug design and parallel synthesis
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
8VLT
 
 | |
4DPF
 
 | | BACE-1 in complex with a HEA-macrocyclic type inhibitor | | Descriptor: | Beta-secretase 1, N-[(4S,8E,11S)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-2,13-dioxo-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraen-16-yl]-N-methylmethanesulfonamide | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
3DM6
 
 | | Beta-secretase 1 complexed with statine-based inhibitor | | Descriptor: | 5-[[(2S)-2-[[(3R,4S)-5-(3,5-difluorophenoxy)-3-hydroxy-4-[[3-(methyl-methylsulfonyl-amino)-5-[[(1R)-1-phenylethyl]carbamoyl]phenyl]carbonylamino]pentanoyl]amino]-3-methyl-butanoyl]amino]benzene-1,3-dicarboxylic acid, Beta-secretase 1, ISOPROPYL ALCOHOL | | Authors: | Lindberg, J, Borkakoti, N, Nystrom, S. | | Deposit date: | 2008-06-30 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and SAR of potent statine-based BACE-1 inhibitors: exploration of P1 phenoxy and benzyloxy residues
Bioorg.Med.Chem., 16, 2008
|
|
4DPI
 
 | | BACE-1 in complex with HEA-macrocyclic inhibitor, MV078512 | | Descriptor: | (4S,8E,11R)-4-[(1R)-1-hydroxy-2-{[3-(propan-2-yl)benzyl]amino}ethyl]-16-methyl-11-phenyl-6-oxa-3,12-diazabicyclo[12.3.1]octadeca-1(18),8,14,16-tetraene-2,13-dione, Beta-secretase 1 | | Authors: | Lindberg, J, Borkakoti, N, Derbyshire, D. | | Deposit date: | 2012-02-13 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent macrocyclic BACE-1 inhibitors incorporating a hydroxyethylamine core: design, synthesis and X-ray crystal structures of enzyme inhibitor complexes.
Bioorg.Med.Chem., 20, 2012
|
|
4EO4
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with seryl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonine--tRNA ligase, mitochondrial, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Soll, D, Simonovic, M. | | Deposit date: | 2012-04-13 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase.
J.Biol.Chem., 287, 2012
|
|
2R2M
 
 | | 2-(2-Chloro-6-Fluorophenyl)Acetamides as Potent Thrombin Inhibitors | | Descriptor: | Hirudin-3A, N-[2-({[amino(imino)methyl]amino}oxy)ethyl]-2-{6-chloro-3-[(2,2-difluoro-2-phenylethyl)amino]-2-fluorophenyl}acetamide, Thrombin heavy chain, ... | | Authors: | Spurlino, J. | | Deposit date: | 2007-08-27 | | Release date: | 2008-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2-(2-Chloro-6-Fluorophenyl)Acetamides as Potent Thrombin Inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3UGT
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase - orthorhombic crystal form | | Descriptor: | Threonyl-tRNA synthetase, mitochondrial, ZINC ION | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1G2K
 
 | | HIV-1 PROTEASE WITH CYCLIC SULFAMIDE INHIBITOR, AHA047 | | Descriptor: | 3-(7-BENZYL-4,5-DIHYDROXY-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1L6-[1,2,7]THIADIAZEPAN-2-YLMETHYL)-N-METHYL-BENZAMIDE, PROTEASE RETROPEPSIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-20 | | Release date: | 2001-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
1G35
 
 | | CRYSTAL STRUCTURE OF HIV-1 PROTEASE IN COMPLEX WITH INHIBITOR, AHA024 | | Descriptor: | 2-[4-(HYDROXY-METHOXY-METHYL)-BENZYL]-7-(4-HYDROXYMETHYL-BENZYL)-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-1LAMBDA6-[1,2,7]THIADIAZEPANE-4,5-DIOL, HIV-1 PROTEASE | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2000-10-23 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and comparative molecular field analysis (CoMFA) of symmetric and nonsymmetric cyclic sulfamide HIV-1 protease inhibitors.
J.Med.Chem., 44, 2001
|
|
3UH0
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with threonyl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UGQ
 
 | | Crystal structure of the apo form of the yeast mitochondrial threonyl-tRNA synthetase determined at 2.1 Angstrom resolution | | Descriptor: | POTASSIUM ION, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4TYB
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TXS
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (4-hydroxyphenyl)acetonitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
3DA6
 
 | | Crystal Structure of human JNK3 complexed with N-(3-methyl-4-(3-(2-(methylamino)pyrimidin-4-yl)pyridin-2-yloxy)naphthalen-1-yl)-1H-benzo[d]imidazol-2-amine | | Descriptor: | Mitogen-activated protein kinase 10, N-[3-methyl-4-({3-[2-(methylamino)pyrimidin-4-yl]pyridin-2-yl}oxy)naphthalen-1-yl]-1H-benzimidazol-2-amine | | Authors: | Cee, V.J, Cheng, A.C, Romero, K, Bellon, S, Mohr, C, Whittington, D.A, Bready, J, Caenepeel, S, Coxon, A, Deak, H.L, Hodous, B.L, Kim, J.L, Lin, J, Nguyen, H, Olivieri, P.R, Patel, V.F, Wang, L, Hughes, P, Geuns-Meyer, S. | | Deposit date: | 2008-05-28 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyridyl-pyrimidine benzimidazole derivatives as potent, selective, and orally bioavailable inhibitors of Tie-2 kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4KFN
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4W2E
 
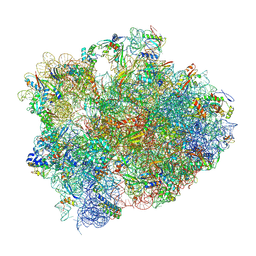 | | Crystal structure of Elongation Factor 4 (EF4/LepA) bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Lin, J, Steitz, T.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-01 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome.
Science, 345, 2014
|
|
4KJZ
 
 | |
6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
