3TP4
 
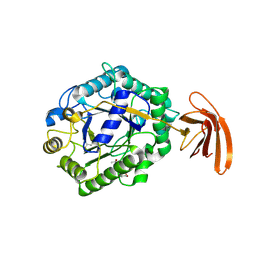 | | Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
6H3M
 
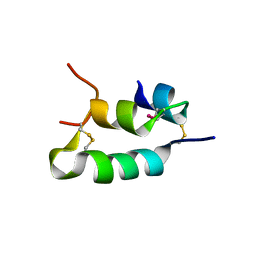 | |
3V45
 
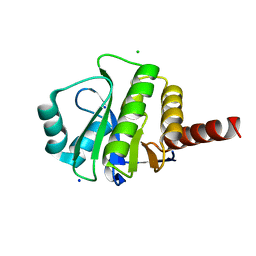 | | Crystal Structure of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium Target OR130 | | Descriptor: | CHLORIDE ION, SODIUM ION, Serine hydrolase OSH55 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JKO
 
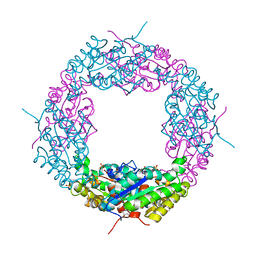 | | Crystal structure of a catalytic mutant of Axe2 (Axe2_S15A), an acetylxylan esterase from Geobacillus stearothermophilus | | Descriptor: | Acetyl xylan esterase, GLYCEROL, PHOSPHATE ION | | Authors: | Azulay, D, Lansky, S, Alalouf, O, Solomon, V, Alhassid, A, Belrahli, H, Govada, L, Chayan, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2013-03-11 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique octameric structure of Axe2, an intracellular acetyl-xylooligosaccharide esterase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5WKO
 
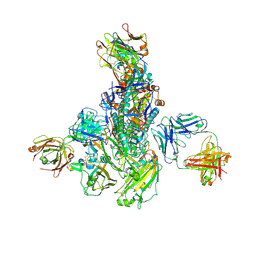 | |
3T03
 
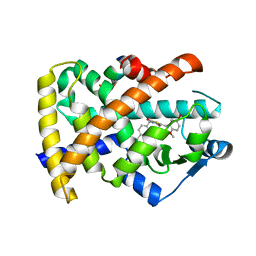 | | Crystal structure of PPAR gamma ligand binding domain in complex with a novel partial agonist GQ-16 | | Descriptor: | (5Z)-5-(5-bromo-2-methoxybenzylidene)-3-(4-methylbenzyl)-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Rajagopalan, S, Webb, P, Baxter, J.D, Brennan, R.G, Phillips, K.J. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
J.Biol.Chem., 287, 2012
|
|
5WKZ
 
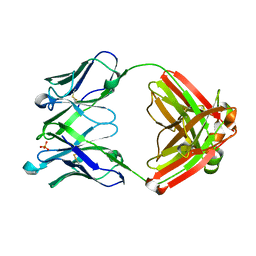 | | VH1-69 germline antibody predicted from CR6261 | | Descriptor: | Immunoglobulin heavy variable 1-69D,IgG H chain, Lambda-chain (AA -20 to 215), SULFATE ION | | Authors: | Lang, S, Lee, P.S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem
To Be Published
|
|
1JYA
 
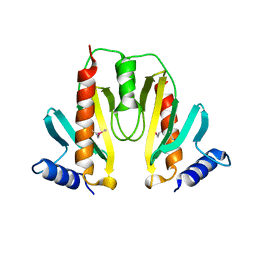 | | Crystal Structure of SycE | | Descriptor: | YOPE regulator | | Authors: | Ghosh, P, Birtalan, S. | | Deposit date: | 2001-09-11 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the Yersinia type III secretory system chaperone SycE.
Nat.Struct.Biol., 8, 2001
|
|
4ESS
 
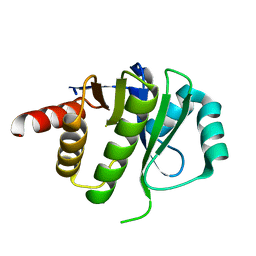 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4EQ7
 
 | | Structure of Atu4243-GABA receptor | | Descriptor: | ABC transporter, substrate binding protein (Polyamine), GLYCEROL, ... | | Authors: | Morera, S, Planamente, S. | | Deposit date: | 2012-04-18 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
4ETK
 
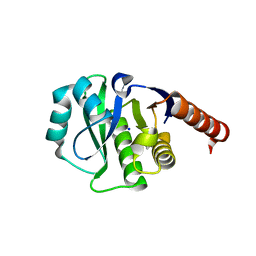 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4F2V
 
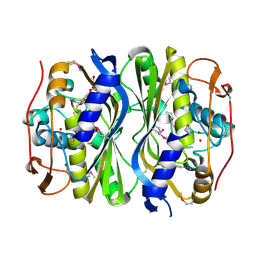 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4DRT
 
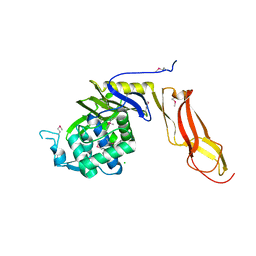 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4EUO
 
 | | Structure of Atu4243-GABA sensor | | Descriptor: | ABC transporter, substrate binding protein (Polyamine), GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Morera, S, Planamente, S. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for selective GABA binding in bacterial pathogens.
Mol.Microbiol., 86, 2012
|
|
4E9D
 
 | | The structure of the polo-box domain (PBD) of polo-like kinase 1 (Plk1) in complex with 3-(1-benzothiophen-2-yl)propanoyl-derivatized DPPLHSpTA peptide | | Descriptor: | 3-(1-benzothiophen-2-yl)propanoyl-derivatized DPPLHSpTA peptide, GLYCEROL, Serine/threonine-protein kinase PLK1 | | Authors: | Sledz, P, Hyvonen, M, Lang, S, Stubbs, C.J, Abell, C. | | Deposit date: | 2012-03-21 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | High-throughput interrogation of ligand binding mode using a fluorescence-based assay.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
5AEQ
 
 | | Neuronal calcium sensor (NCS-1)from Rattus norvegicus | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, SODIUM ION | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5AER
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with D2 dopamine receptor peptide from Homo sapiens | | Descriptor: | CALCIUM ION, D(2) DOPAMINE RECEPTOR, NEURONAL CALCIUM SENSOR 1, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5AFP
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with rhodopsin kinase peptide from Homo sapiens | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, RHODOPSIN KINASE, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
4D0R
 
 | | Mtb InhA complex with Pyradizinone compound 1 | | Descriptor: | 4-(4-chlorophenoxy)-6-oxo-1-phenyl-1,6-dihydropyridazine-3-carboxamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Lange, S. | | Deposit date: | 2014-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyridazinones: A Novel Scaffold with Excellent Physicochemical Properties and Safety Profile for a Clinically Validated Target of Mycobacterium Tuberculosis
To be Published
|
|
3MWP
 
 | | Nucleoprotein structure of lassa fever virus | | Descriptor: | Nucleoprotein, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MX5
 
 | | Lassa fever virus nucleoprotein complexed with UTP | | Descriptor: | Nucleoprotein, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MWT
 
 | | Crystal structure of Lassa fever virus nucleoprotein in complex with Mn2+ | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
3MX2
 
 | | Lassa fever virus Nucleoprotein complexed with dTTP | | Descriptor: | Nucleoprotein, THYMIDINE-5'-TRIPHOSPHATE, ZINC ION | | Authors: | Qi, X, Lan, S, Wang, W, Schelde, L.M, Dong, H, Wallat, G, Liang, Y, Ly, H, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2010-05-06 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Nature, 468, 2010
|
|
1KK0
 
 | |
