7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | Descriptor: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8W
 
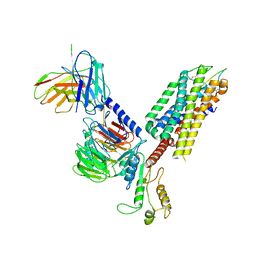 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gq | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8V
 
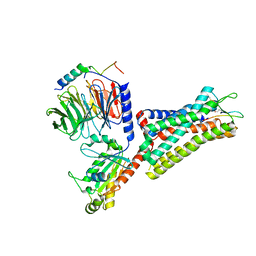 | | Cryo-EM structure of the cholecystokinin receptor CCKBR in complex with gastrin-17 and Gi | | Descriptor: | Gastrin-17, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F58
 
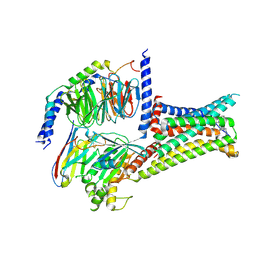 | | Cryo-EM structure of THIQ-MC4R-Gs_Nb35 complex | | Descriptor: | (3R)-N-[(2R)-3-(4-chlorophenyl)-1-[4-cyclohexyl-4-(1,2,4-triazol-1-ylmethyl)piperidin-1-yl]-1-oxidanylidene-propan-2-yl]-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F54
 
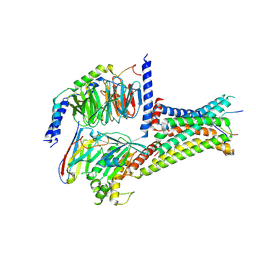 | | Cryo-EM structure of afamelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F55
 
 | | Cryo-EM structure of bremelanotide-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7F53
 
 | | Cryo-EM structure of a-MSH-MC4R-Gs_Nb35 complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, L, Mao, C, Shen, Q, Yang, D, Shen, D, Qin, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into ligand recognition and activation of the melanocortin-4 receptor.
Cell Res., 31, 2021
|
|
7FAS
 
 | | VAR2CSA 3D7 ectodomain core region | | Descriptor: | Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Zhaoning, W. | | Deposit date: | 2021-07-07 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
7F8X
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with NN9056 | | Descriptor: | ASP-SMF-NLE-GLY-TRP-NLE-OEM-MEA-NH2 (NN9056), Cholecystokinin receptor type A,Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7FAP
 
 | | Structure of VAR2CSA-CSA 3D7 | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-3)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, Erythrocyte membrane protein 1, PfEMP1 | | Authors: | Wang, L, Wang, Z. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The molecular mechanism of cytoadherence to placental or tumor cells through VAR2CSA from Plasmodium falciparum.
Cell Discov, 7, 2021
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMR
 
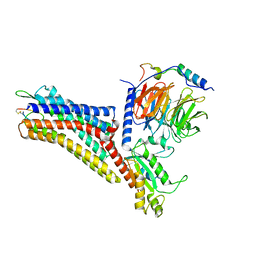 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
3CUV
 
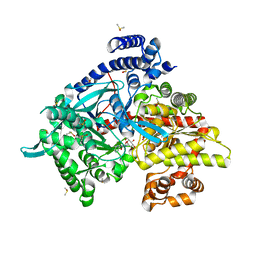 | | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
3CUW
 
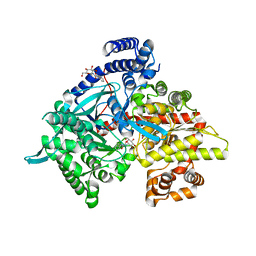 | | Crystal structure of glycogen phosphorylase b in complex with N-(-D-glucopyranosyl)-N'-(2-naphthyl)oxamides | | Descriptor: | Glycogen phosphorylase, muscle form, N-[oxo(phenylamino)acetyl]-beta-D-glucopyranosylamine | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
3CUU
 
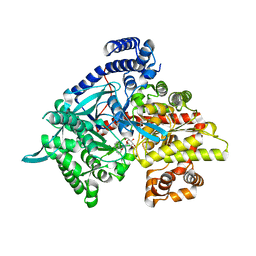 | | Crystal structure of glycogen phosphorylase b in complex with N-(-D-glucopyranosyl)-N'-(2-naphthyl)oxamides | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(naphthalen-1-ylamino)(oxo)acetyl]-beta-D-glucopyranosylamine | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
3CUT
 
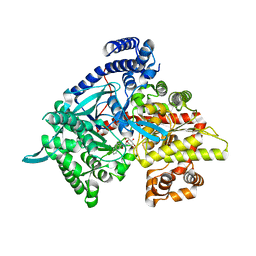 | | Crystal structure of glycogen phosphorylase b in complex with N-(-D-glucopyranosyl)-N'-(2-naphthyl)oxamide | | Descriptor: | Glycogen phosphorylase, muscle form, N-[(naphthalen-2-ylamino)(oxo)acetyl]-beta-D-glucopyranosylamine | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
7RR0
 
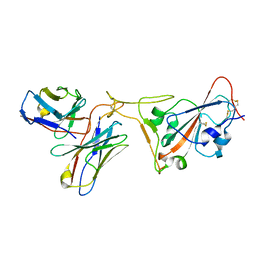 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 222 Fab Heavy Chain, PDI 222 Fab Light Chain, ... | | Authors: | Pymm, P, Glukhova, A, Black, K.A, Tham, W.H. | | Deposit date: | 2021-08-08 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7U2K
 
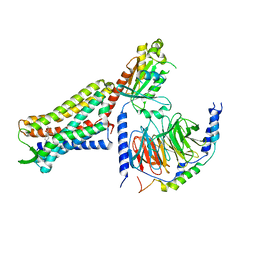 | | C6-guano bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based design of bitopic ligands for the μ-opioid receptor.
Nature, 613, 2023
|
|
7MZK
 
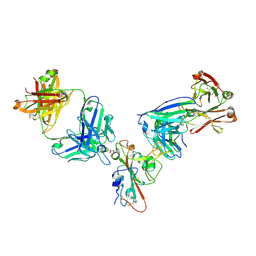 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 96 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZH
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 119 | | Descriptor: | Spike protein S1, WCSL 119 heavy chain, WCSL 119 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZG
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PDI 42 heavy chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZJ
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 93 | | Descriptor: | GLYCEROL, PDI 93 heavy chain, PDI 93 light chain, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
