6V9N
 
 | |
5DFN
 
 | | Structure of Tetrahymena Telomerase P45 C-terminal domain | | Descriptor: | Telomerase associated protein p45 | | Authors: | Chan, H, Cascio, D, Sawaya, M.R, Feigon, J. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions.
Science, 350, 2015
|
|
6TUK
 
 | | Crystal structure of Fdr9 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification and crystal structure determination of a ferredoxin reductase from the actinobacterium Thermobifida fusca.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TKA
 
 | | Crystal structure of human O-GlcNAc transferase bound to substrate 7 and a peptide from HCF-1 pro-repeat 2 (11-26) | | Descriptor: | 3-[2,2-bis(fluoranyl)-10,12-dimethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9,11-tetraen-4-yl]-~{N}-ethyl-propanamide, HCF-1 pro-repeat 2 (11-26), UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A Direct Fluorescent Activity Assay for Glycosyltransferases Enables Convenient High-Throughput Screening: Application to O-GlcNAc Transferase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5E8J
 
 | |
5E8Y
 
 | |
6UWQ
 
 | |
6V1G
 
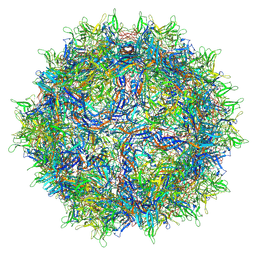 | | Genome-containing AAVrh.10 | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6UYB
 
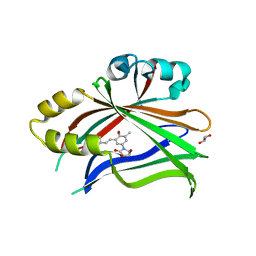 | | Crystal structure of TEAD2 bound to Compound 1 | | Descriptor: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
6V1T
 
 | | Empty AAVrh.39 particle | | Descriptor: | Capsid protein VP1 | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-21 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6V6A
 
 | | Inhibitory scaffolding of the ancient MAPK, ERK7 | | Descriptor: | 1,2-ETHANEDIOL, Apical Cap Protein 9 (AC9), Mitogen-activated protein kinase | | Authors: | Dewangan, P.S, O'Shaughnessy, W.J, Back, P.S, Hu, X, Bradley, P.J, Reese, M.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ancient MAPK ERK7 is regulated by an unusual inhibitory scaffold required forToxoplasmaapical complex biogenesis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VFH
 
 | |
5CWN
 
 | |
5D6H
 
 | | Crystal structure of CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Csu biofilm-mediating pili of Acinetobacter baumannii | | Descriptor: | CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-04 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis.
Plos Pathog., 11, 2015
|
|
6TNR
 
 | | PI3K delta in complex with N[5(7{2[4(2hydroxypropan2yl)piperidin1 yl]ethoxy}1,3dihydro2benzofuran5yl)2 methoxypyridin3yl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[2-methoxy-5-[7-[2-[4-(2-oxidanylpropan-2-yl)piperidin-1-yl]ethoxy]-1,3-dihydro-2-benzofuran-5-yl]pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Rowland, P, Henley, Z.A, Barton, N, Down, K. | | Deposit date: | 2019-12-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of Orally Bioavailable PI3K delta Inhibitors and Identification of Vps34 as a Key Selectivity Target.
J.Med.Chem., 63, 2020
|
|
6SAR
 
 | | E coli BepA/YfgC | | Descriptor: | Beta-barrel assembly-enhancing protease, SULFATE ION, ZINC ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Function Characterization of the Conserved Regulatory Mechanism of the Escherichia coli M48 Metalloprotease BepA.
J.Bacteriol., 203, 2020
|
|
6V0P
 
 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
6VFI
 
 | |
6UWW
 
 | |
5BVB
 
 | | Engineered Digoxigenin binder DIG5.1a | | Descriptor: | DIG5.1a, DIGOXIGENIN | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2015-06-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | CSAR Benchmark Exercise 2013: Evaluation of Results from a Combined Computational Protein Design, Docking, and Scoring/Ranking Challenge.
J.Chem.Inf.Model., 56, 2016
|
|
6UVP
 
 | | BACE-1 in complex with compound #3 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]cyclopropyl}-5-fluoropyridine-2-carboxamide, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UWV
 
 | | BACE-1 in complex with compound #34 | | Descriptor: | (4aR,7aR)-7a-[(1R,2R)-2-(2-{[(1R,2R)-2-methylcyclopropyl]methoxy}propan-2-yl)cyclopropyl]-6-(pyrimidin-2-yl)-4,4a,5,6,7,7a-hexahydropyrrolo[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Stout, S.L. | | Deposit date: | 2019-11-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UYC
 
 | | Crystal structure of TEAD2 bound to Compound 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-{5-[(E)-2-(4,4-difluorocyclohexyl)ethenyl]-6-methoxypyridin-3-yl}methanesulfonamide, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
6UVY
 
 | | BACE-1 in complex with compound #18 | | Descriptor: | (1R,2R)-2-[(4aS,7aR)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-N-{[(1R,2R)-2-methylcyclopropyl]methyl}cyclopropane-1-carboxamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Hendle, J, Timm, D.E. | | Deposit date: | 2019-11-04 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Preparation and biological evaluation of BACE1 inhibitors: Leveraging trans-cyclopropyl moieties as ligand efficient conformational constraints.
Bioorg.Med.Chem., 28, 2020
|
|
6UZ1
 
 | |
