2QXW
 
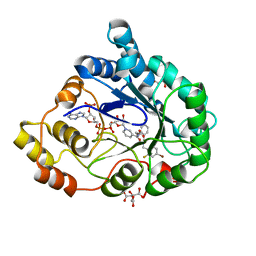 | | Perdeuterated alr2 in complex with idd594 | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O, Cousido-Siah, A, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1MKM
 
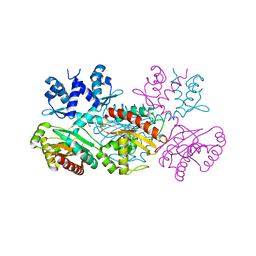 | | CRYSTAL STRUCTURE OF THE THERMOTOGA MARITIMA ICLR | | Descriptor: | FORMIC ACID, IclR transcriptional regulator, ZINC ION | | Authors: | Kim, Y, Zhang, R.G, Joachimiak, A, Skarina, T, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Thermotoga maritima 0065, a member of the IclR transcriptional factor family.
J.Biol.Chem., 277, 2002
|
|
2K4C
 
 | | tRNAPhe-based homology model for tRNAVal refined against base N-H RDCs in two media and SAXS data | | Descriptor: | 76-MER | | Authors: | Grishaev, A, Ying, J, Canny, M.D, Pardi, A, Bax, A. | | Deposit date: | 2008-06-04 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data.
J.Biomol.Nmr, 42, 2008
|
|
1MKZ
 
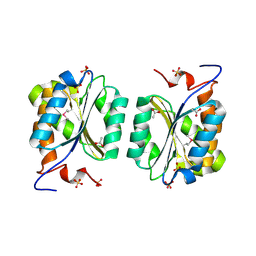 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
1W82
 
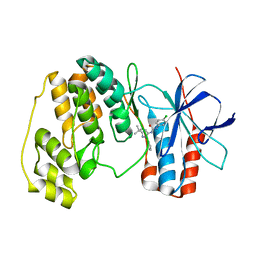 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, N-[(3Z)-5-TERT-BUTYL-2-PHENYL-1,2-DIHYDRO-3H-PYRAZOL-3-YLIDENE]-N'-(4-CHLOROPHENYL)UREA | | Authors: | Tickle, J, Jhoti, H, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-16 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation
J.Med.Chem., 48, 2005
|
|
2KZR
 
 | | Solution NMR Structure of Ubiquitin thioesterase OTU1 (EC 3.1.2.-) from Mus musculus, Northeast Structural Genomics Consortium Target MmT2A | | Descriptor: | Ubiquitin thioesterase OTU1 | | Authors: | Chitayat, S, Gutmanas, A, Lemak, A, Yee, A, Bezsonova, I, Wu, B, Doherty, R.S, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target MmT2A
To be Published
|
|
2LF0
 
 | | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering, NESG target SfR339/OCSP target sf3636 | | Descriptor: | Uncharacterized protein yibL | | Authors: | Wu, B, Lemak, A, Yee, A, Lee, H, Gutmanas, A, Semesi, A, Garcia, M, Fang, X, Wang, Y, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering
To be Published
|
|
2FAM
 
 | | X-RAY CRYSTAL STRUCTURE OF FERRIC APLYSIA LIMACINA MYOGLOBIN IN DIFFERENT LIGANDED STATES | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, THIOCYANATE ION | | Authors: | Conti, E, Moser, C, Rizzi, M, Mattevi, A, Lionetti, C, Coda, A, Ascenzi, P, Brunori, M, Bolognesi, M. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of ferric Aplysia limacina myoglobin in different liganded states.
J.Mol.Biol., 233, 1993
|
|
2FAL
 
 | | X-RAY CRYSTAL STRUCTURE OF FERRIC APLYSIA LIMACINA MYOGLOBIN IN DIFFERENT LIGANDED STATES | | Descriptor: | CYANIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Conti, E, Moser, C, Rizzi, M, Mattevi, A, Lionetti, C, Coda, A, Ascenzi, P, Brunori, M, Bolognesi, M. | | Deposit date: | 1993-06-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of ferric Aplysia limacina myoglobin in different liganded states.
J.Mol.Biol., 233, 1993
|
|
2LF3
 
 | | Solution NMR structure of HopPmaL_281_385 from Pseudomonas syringae pv. maculicola str. ES4326, Midwest Center for Structural Genomics target APC40104.5 and Northeast Structural Genomics Consortium target PsT2A | | Descriptor: | Effector protein hopAB3 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
7OND
 
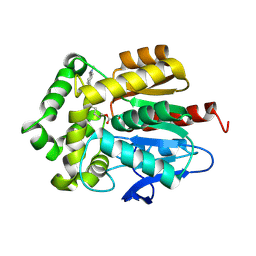 | |
4C90
 
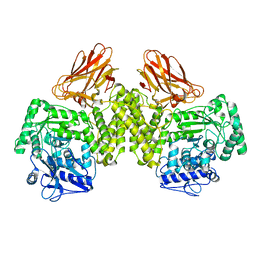 | | Evidence that GH115 alpha-glucuronidase activity is dependent on conformational flexibility | | Descriptor: | ALPHA-GLUCURONIDASE GH115, SODIUM ION | | Authors: | Rogowski, A, Basle, A, Farinas, C.S, Solovyova, A, Mortimer, J.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evidence that Gh115 Alpha-Glucuronidase Activity is Dependent on Conformational Flexibility
J.Biol.Chem., 289, 2014
|
|
7YC6
 
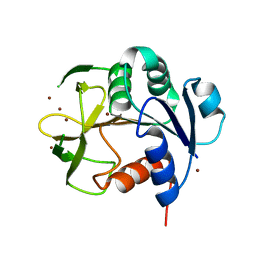 | |
2YAV
 
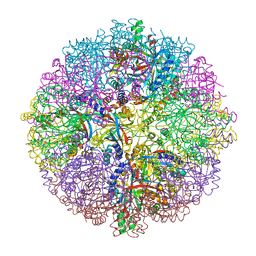 | | ZN INHIBITED SULFUR OXYGENASE REDUCTASE | | Descriptor: | ACETATE ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Veith, A, Urich, T, Seyfarth, K, Protze, J, Frazao, C, Kletzin, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Substrate Pathways and Mechanisms of Inhibition in the Sulfur Oxygenase Reductase of Acidianus Ambivalens.
Front.Microbiol., 2, 2011
|
|
4CDD
 
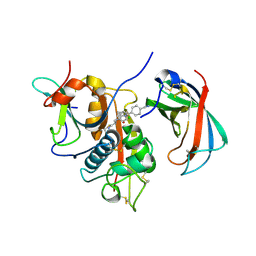 | | Human DPP1 in complex with (2S)-N-((1S)-1-cyano-2-(4-(4-cyanophenyl) phenyl)ethyl)piperidine-2-carboxamide | | Descriptor: | (2S)-N-[(2S)-1-AZANYLIDENE-3-[4-(4-CYANOPHENYL)PHENYL]PROPAN-2-YL]PIPERIDINE-2-CARBOXAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debreczeni, J, Edman, K, Furber, M, Tiden, A, Gardiner, P, Mete, T, Ford, R, Millichip, I, Stein, L, Mather, A, Kinchin, E, Luckhurst, C, Cage, P, Sanghanee, H, Breed, J, Wissler, L. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Cathepsin C Inhibitors: Property Optimization and Identification of a Clinical Candidate.
J.Med.Chem., 57, 2014
|
|
7OO4
 
 | |
5X7S
 
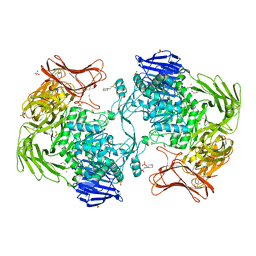 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
4CDE
 
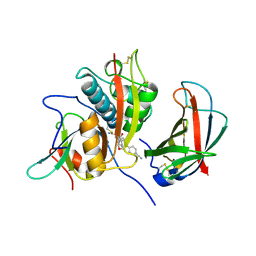 | | Human DPP1 in complex with 4-amino-N-((1S)-1-cyano-2-(4-(4- cyanophenyl)phenyl)ethyl)tetrahydropyran-4-carboxamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AZANYL-N-[(2S)-1-AZANYLIDENE-3-[4-(4-CYANOPHENYL)PHENYL]PROPAN-2-YL]OXANE-4-CARBOXAMIDE, CHLORIDE ION, ... | | Authors: | Debreczeni, J, Edman, K, Furber, M, Tiden, A, Gardiner, P, Mete, T, Ford, R, Millichip, I, Stein, L, Mather, A, Kinchin, E, Luckhurst, C, Cage, P, Sanghanee, H, Breed, J, Wissler, L. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cathepsin C Inhibitors: Property Optimization and Identification of a Clinical Candidate.
J.Med.Chem., 57, 2014
|
|
5XAN
 
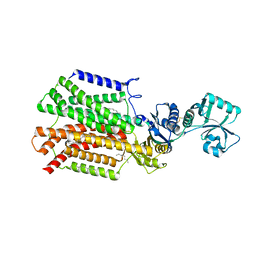 | | Crystal structure of SecDF in I form (P212121 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, POLYETHYLENE GLYCOL (N=34), Protein translocase subunit SecD | | Authors: | Tsukazaki, T, Tanaka, Y, Furukwa, A. | | Deposit date: | 2017-03-14 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Tunnel Formation Inferred from the I-Form Structures of the Proton-Driven Protein Secretion Motor SecDF
Cell Rep, 19, 2017
|
|
5XBP
 
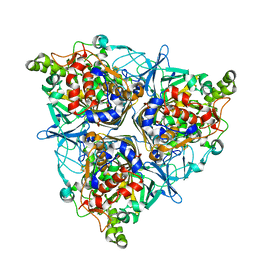 | | Oxygenase component of 3-nitrotoluene dioxygenase from Diaphorobacter sp. strain DS2 | | Descriptor: | 3NT oxygenase alpha subunit, 3NT oxygenase beta subunit, FE (III) ION, ... | | Authors: | Ramaswamy, S, Kumari, A, Singh, D, Gurunath, R. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional studies of ferredoxin and oxygenase components of 3-nitrotoluene dioxygenase from Diaphorobacter sp. strain DS2.
PLoS ONE, 12, 2017
|
|
7OSD
 
 | |
7OS8
 
 | | NMR SOLUTION STRUCTURE OF [Pro3,DLeu9]TL | | Descriptor: | PHE-VAL-PRO-TRP-PHE-SER-LYS-PHE-DLE-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-11 | | Last modified: | 2021-08-25 | | Method: | SOLUTION NMR | | Cite: | First-in-Class Cyclic Temporin L Analogue: Design, Synthesis, and Antimicrobial Assessment.
J.Med.Chem., 64, 2021
|
|
5R4M
 
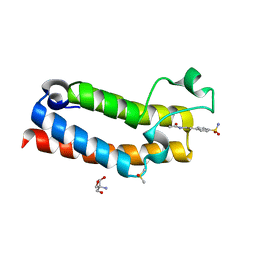 | | PanDDA analysis group deposition -- CRYSTAL STRUCTURE OF THE BROMODOMAIN OF HUMAN NUCLEOSOME-REMODELING FACTOR SUBUNIT BPTF in complex with FMOPL000513a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, N-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}ACETAMIDE, ... | | Authors: | Talon, R, Krojer, T, Fairhead, M, Sethi, R, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Wright, N, MacLean, E, Renjie, Z, Dias, A, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2020-02-24 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8UTK
 
 | | IL-23R minibinder - 23R-B04dslf02IB | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 23R-B04dslf02IB, ... | | Authors: | Bera, A.K, Berger, S.A, Kang, A, Baker, D. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Preclinical proof of principle for orally delivered Th17 antagonist miniproteins.
Cell, 2024
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
