7YZ6
 
 | | Molecular snapshots of drug release from tubulin: Dark (steady state) | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YZ3
 
 | | Molecular snapshots of drug release from tubulin: Apo state | | Descriptor: | CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YZ1
 
 | | Molecular snapshots of drug release from tubulin: 1 millisecond after photoactivation. | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
7YYY
 
 | | Molecular snapshots of drug release from tubulin: 1 microsecond after photoactivation | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Wranik, M, Weinert, T, Standfuss, J. | | Deposit date: | 2022-02-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Watching the release of a photopharmacological drug from tubulin using time-resolved serial crystallography.
Nat Commun, 14, 2023
|
|
6RYP
 
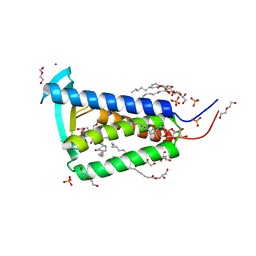 | | Bacterial membrane enzyme structure by the in meso method at 2.3 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Lipoprotein signal peptidase, ... | | Authors: | Huang, C.Y, Olatunji, S, Bailey, J, Yu, X, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2019-06-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of lipoprotein signal peptidase II from Staphylococcus aureus complexed with antibiotics globomycin and myxovirescin.
Nat Commun, 11, 2020
|
|
6K2N
 
 | |
3BYZ
 
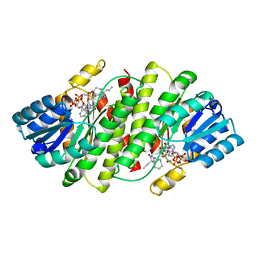 | | 2-Amino-1,3-thiazol-4(5H)-ones as Potent and Selective 11-Hydroxysteroid Dehydrogenase Type 1 Inhibitors | | Descriptor: | (5S)-2-(cyclooctylamino)-5-methyl-5-propyl-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | 2-amino-1,3-thiazol-4(5H)-ones as potent and selective 11beta-hydroxysteroid dehydrogenase type 1 inhibitors: enzyme-ligand co-crystal structure and demonstration of pharmacodynamic effects in C57Bl/6 mice.
J.Med.Chem., 51, 2008
|
|
6KBW
 
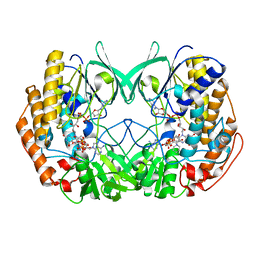 | | Crystal structure of Tmm from Myroides profundi D25 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethylamine monooxygenase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Oxidation of trimethylamine to trimethylamine N-oxide facilitates high hydrostatic pressure tolerance in a generalist bacterial lineage.
Sci Adv, 7, 2021
|
|
8SPH
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
8SPI
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
7KSO
 
 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KSR
 
 | | PRC2:EZH1_A from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KTP
 
 | | PRC2:EZH1_B from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
9ERY
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-[bis(fluoranyl)methyl]-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]methoxy]-2-[(2-hydroxyethylamino)methyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Plewka, J, Magiera-Mularz, K, Zhang, W. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of antitumor activity of 2-arylmethoxy-4-(2-fluoromethyl-biphenyl-3-ylmethoxy) benzylamine derivatives as PD-1/PD-l1 inhibitors.
Eur.J.Med.Chem., 276, 2024
|
|
2RBE
 
 | | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors | | Descriptor: | (5R)-2-[(2-fluorophenyl)amino]-5-(1-methylethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2007-09-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8TXT
 
 | | Crystal structure of 05.GC.w13.02 Fab in complex with H5 HA from A/Viet Nam/1203/2004(H5N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GC_W13B_B, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
7RQF
 
 | |
7RQH
 
 | |
7RPQ
 
 | |
8DNN
 
 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
8H8C
 
 | | Type VI secretion system effector RhsP in its post-autoproteolysis and dimeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8B
 
 | | Type VI secretion system effector RhsP in its pre-autoproteolysis and monomeric form | | Descriptor: | Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
8H8A
 
 | | Type VI secretion system effector RhsP in its post-autoproteolysis and monomeric form | | Descriptor: | C-terminal peptide from Putative Rhs-family protein, Putative Rhs-family protein | | Authors: | Tang, L, Dong, S.Q, Rasheed, N, Wu, H.W, Zhou, N.K, Li, H.D, Wang, M.L, Zheng, J, He, J, Chao, W.C.H. | | Deposit date: | 2022-10-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Vibrio parahaemolyticus prey targeting requires autoproteolysis-triggered dimerization of the type VI secretion system effector RhsP.
Cell Rep, 41, 2022
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
