2Y40
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
1LYN
 
 | |
1LVL
 
 | |
6ZEZ
 
 | |
1MZN
 
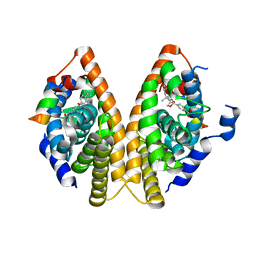 | | CRYSTAL STRUCTURE at 1.9 ANGSTROEMS RESOLUTION OF THE HOMODIMER OF HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO THE SYNTHETIC AGONIST COMPOUND BMS 649 AND A COACTIVATOR PEPTIDE | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-10-09 | | Release date: | 2002-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
8DVC
 
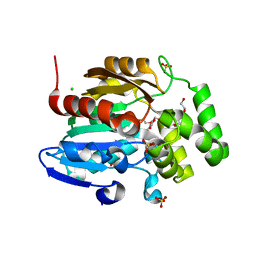 | | Receptor ShHTL5 from Striga hermonthica in complex with strigolactone agonist GR24 | | Descriptor: | (3R,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methyl)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Arellano-Saab, A, Skarina, T, Yim, V, Savchenko, A, Stogios, P.J, McCourt, P. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | Structural analysis of a hormone-bound Striga strigolactone receptor.
Nat.Plants, 9, 2023
|
|
7R60
 
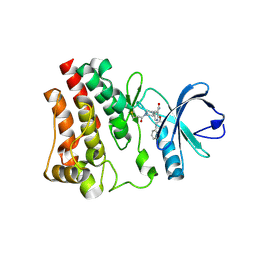 | | BTK in complex with 18A | | Descriptor: | 2-(4-phenoxyphenoxy)-5-[(3R)-1-(prop-2-enoyl)piperidin-3-yl]pyridine-3-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Covalent Bruton's Tyrosine Kinase Inhibitors with Decreased CYP2C8 Inhibitory Activity.
Chemmedchem, 16, 2021
|
|
7R61
 
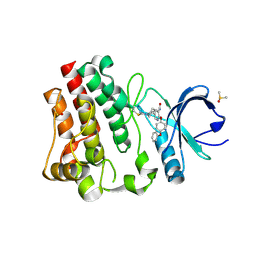 | | BTK in complex with 25A | | Descriptor: | 5-{(3S)-1-[(Z)-iminomethyl]piperidin-3-yl}-2-(4-phenoxyphenoxy)pyridine-3-carboxamide, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A. | | Deposit date: | 2021-06-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of Covalent Bruton's Tyrosine Kinase Inhibitors with Decreased CYP2C8 Inhibitory Activity.
Chemmedchem, 16, 2021
|
|
1N0V
 
 | | Crystal structure of elongation factor 2 | | Descriptor: | Elongation factor 2 | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
1N1Y
 
 | | Trypanosoma rangeli sialidase in complex with sialic acid | | Descriptor: | N-acetyl-alpha-neuraminic acid, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-21 | | Release date: | 2003-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution structures of free and inhibitor-bound
Trypanosoma rangeli sialidase and its comparison with T.
cruzi trans-sialidase
J.Mol.Biol., 325, 2003
|
|
1N1T
 
 | | Trypanosoma rangeli sialidase in complex with DANA at 1.6 A | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SULFATE ION, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-20 | | Release date: | 2003-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structures of free and
inhibitor-bound Trypanosoma rangeli
sialidase and its comparison with T. cruzi
trans-sialidase
J.Mol.Biol., 325, 2003
|
|
4OO4
 
 | |
8DWR
 
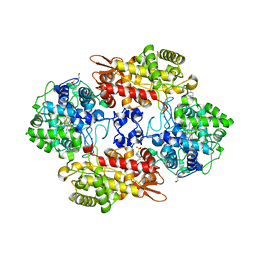 | | Crystal structure of the L333V variant of catalase-peroxidase from Mycobacterium tuberculosis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Diaz-Vilchis, A, Uribe-Vazquez, B, Avila-Linares, A, Rudino-Pinera, E, Soberon, X. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of a catalase-peroxidase variant (L333V-KatG) identified in an INH-resistant Mycobacterium tuberculosis clinical isolate.
Biochem Biophys Rep, 37, 2024
|
|
7RD1
 
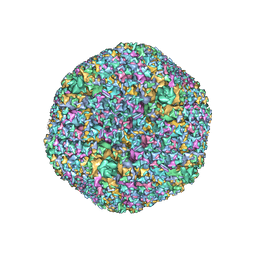 | | The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25 | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Baker, A.T, Boyd, R.J, Sarkar, D, Vermaas, J.V, Williams, D, Singharoy, A. | | Deposit date: | 2021-07-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome.
Sci Adv, 7, 2021
|
|
4UII
 
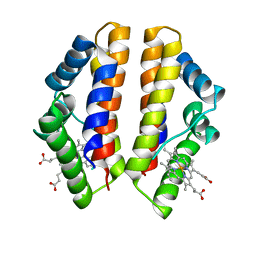 | | Crystal structure of the Azotobacter vinelandii globin-coupled oxygen sensor in the aquo-met form | | Descriptor: | GGDEF DOMAIN PROTEIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Germani, F, De Schutter, A, Pesce, A, Berghmans, H, Van Hauwaert, M.-L, Cuypers, B, Bruno, S, Mozzarelli, A, Moens, L, Van Doorslaer, S, Bolognesi, M, Nardini, M, Dewilde, S. | | Deposit date: | 2015-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.827 Å) | | Cite: | Azotobacter Vinelandii Globin-Coupled Oxygen Sensor is a Diguanylate Cyclase with a Biphasic Oxygen Dissociation
To be Published
|
|
4UNK
 
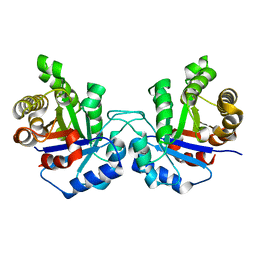 | | Crystal structure of human triosephosphate isomerase (mutant N15D) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Triosephosphate Isomerase (Mutant N15D)
To be Published
|
|
4UNL
 
 | | Crystal structure of a single mutant (N71D) of triosephosphate isomerase from human | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | DeLaMora-DeLaMora, I, Torres-Larios, A, Enriquez-Flores, S, Mendez, S.T, Castillo-Villanueva, A, Gomez-Manzo, S, Lopez-Velazquez, G, Marcial-Quino, J, Torres-Arroyo, A, Garcia-Torres, I, Reyes-Vivas, H, Oria-Hernandez, J. | | Deposit date: | 2014-05-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Single Mutant (N71D) of Triosephosphate Isomerase from Human
To be Published
|
|
4UV8
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Benzyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4S)-5-[(4aS,10aS)-4a-[(1S)-3-azanylidene-1,4-diphenyl-butyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-5,10a-dihydro-1H-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentoxy]-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4V25
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-chloro-5-methylpyrimidin-4-yl)phenyl]-N-(4-{[(difluoroacetyl)amino]methyl}benzyl)-2,4-dihydroxybenzamide, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4V26
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-CHLORO-5-METHYLPYRIMIDIN-4-YL)PHENYL]-2,4-DIHYDROXY-N-(4-{[(TRIFLUOROACETYL)AMINO]METHYL}BENZYL)BENZAMIDE, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
3TMK
 
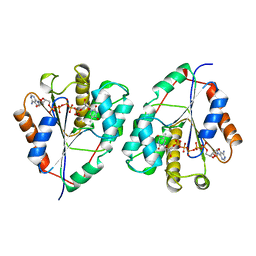 | | CRYSTAL STRUCTURE OF YEAST THYMIDYLATE KINASE COMPLEXED WITH THE BISUBSTRATE INHIBITOR TP5A AT 2.0 A RESOLUTION: IMPLICATIONS FOR CATALYSIS AND AZT ACTIVATION | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDYLATE KINASE | | Authors: | Lavie, A, Schlichting, I, Konrad, M, Goody, R.S, Brundiers, R, Reinstein, J. | | Deposit date: | 1998-01-26 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5'-adenosyl) P5-(5'-thymidyl) pentaphosphate (TP5A) at 2.0 A resolution: implications for catalysis and AZT activation.
Biochemistry, 37, 1998
|
|
6KMK
 
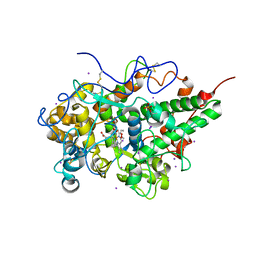 | | Crystal structure of hydrogen peroxide bound bovine lactoperoxidase at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Singh, P.K, Sirohi, H.V, Bhusan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of hydrogen peroxide bound bovine lactoperoxidase at 2.3 A resolution
To Be Published
|
|
3UF1
 
 | | Crystal Structure of Vimentin (fragment 144-251) from Homo sapiens, Northeast Structural Genomics Consortium Target HR4796B | | Descriptor: | Vimentin | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography.
J.Biol.Chem., 287, 2012
|
|
3GP9
 
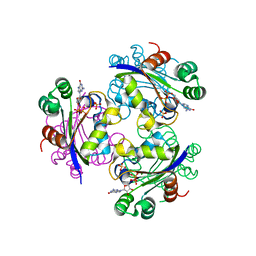 | | Crystal structure of the Mimivirus NDK complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase, ... | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2009-03-23 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3H5J
 
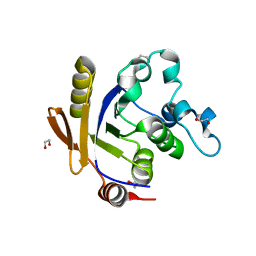 | | LeuD_1-168 small subunit of isopropylmalate isomerase (Rv2987c) from mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydratase small subunit, SULFATE ION | | Authors: | Manikandan, K, Geerlof, A, Weiss, M.S. | | Deposit date: | 2009-04-22 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural studies on the enzyme complex isopropylmalate isomerase (LeuCD) from Mycobacterium tuberculosis
Proteins, 79, 2011
|
|
