4LWO
 
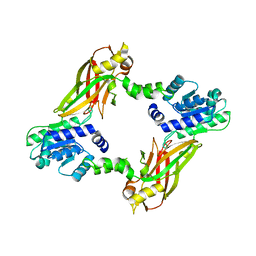 | | Crystal structure of PRMT6 | | Descriptor: | Arginine N-methyltransferase, putative | | Authors: | Zhu, Y, Wang, C, Shi, Y, Teng, M. | | Deposit date: | 2013-07-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Arginine Methyltransferase 6 from Trypanosoma brucei
Plos One, 9, 2014
|
|
4MEJ
 
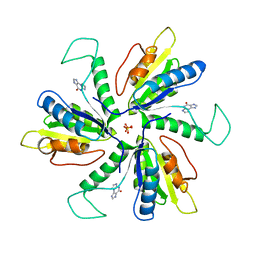 | | Crystal structure of Lactobacillus helveticus purine deoxyribosyl transferase (PDT) with the tricyclic purine 8,9-dihydro-9-oxoimidazo[2,1-b]purine (N2,3-ethenoguanine) | | Descriptor: | 3H-imidazo[2,1-b]purin-4(5H)-one, Nucleoside deoxyribosyltransferase, SULFATE ION | | Authors: | Paul, D, Seckute, J, Ealick, S.E. | | Deposit date: | 2013-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosylation of a tricyclic purine analog at alternative
sites by nucleoside 2 -deoxyribosyltransferases
Plos One, 2014
|
|
4ZSE
 
 | |
4N6J
 
 | | Crystal structure of human Striatin-3 coiled coil domain | | Descriptor: | Striatin-3 | | Authors: | Chen, C, Shi, Z, Zhou, Z. | | Deposit date: | 2013-10-13 | | Release date: | 2014-02-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex.
J.Biol.Chem., 289, 2014
|
|
5BTU
 
 | |
6V03
 
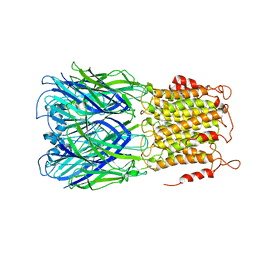 | | ELIC-propylammonium complex in POPC-only nanodiscs | | Descriptor: | 3-AMINOPROPANE, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V0B
 
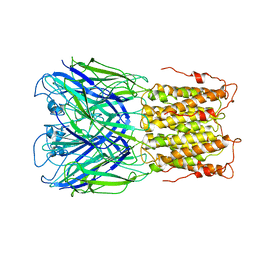 | | Unliganded ELIC in POPC-only nanodiscs. | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Grosman, C, Kumar, P. | | Deposit date: | 2019-11-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7PGO
 
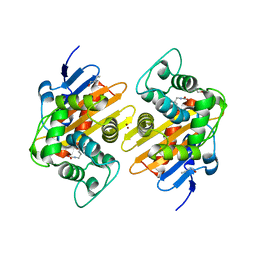 | | Crystal Structure of a Class D Carbapenemase_R250A | | Descriptor: | 1-BUTANOL, BROMIDE ION, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7PFN
 
 | |
7PEH
 
 | | Crystal Structure of a Class D Carbapenemase | | Descriptor: | 1-BUTANOL, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
5BU3
 
 | | Crystal Structure of Diels-Alderase PyrI4 in complex with its product | | Descriptor: | (4S,4aS,6aS,8R,9R,10aR,13R,14aS,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,5,6,6a,7,8,9,10,10a,13,14,18a,18b-hexadecahydro-1H-14a,17-(metheno)benzo[b]naphtho[2,1-h]azacyclododecine-16,18(15H,17H)-dione, GLYCEROL, PyrI4 | | Authors: | Pan, L, Guo, Y, Liu, J. | | Deposit date: | 2015-06-03 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Enzyme-Dependent [4 + 2] Cycloaddition Depends on Lid-like Interaction of the N-Terminal Sequence with the Catalytic Core in PyrI4
Cell Chem Biol, 23, 2016
|
|
2L89
 
 | |
8H08
 
 | |
8H07
 
 | |
6ITP
 
 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6ITQ
 
 | | Crystal structure of cortisol complexed with its nanobody at pH 10.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, SULFATE ION, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
6NVH
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVJ
 
 | | FGFR4 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-fluorophenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-fluorophenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVL
 
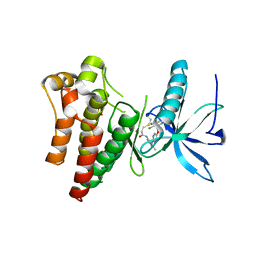 | | FGFR1 complex with N-(2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)-3-methylphenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 1, N-[2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)-3-methylphenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVG
 
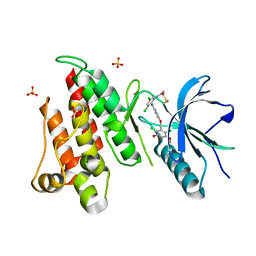 | | FGFR4 complex with N-(3,5-dichloro-2-((5-((2,6-dichloro-3,5-dimethoxybenzyl)oxy)pyrimidin-2-yl)amino)phenyl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[3,5-dichloro-2-({5-[(2,6-dichloro-3,5-dimethoxyphenyl)methoxy]pyrimidin-2-yl}amino)phenyl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
6NVK
 
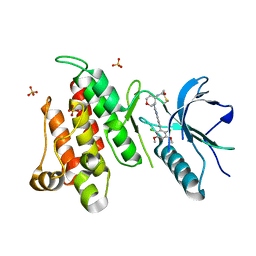 | | FGFR4 complex with BLU-554, N-((3S,4S)-3-((6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl)amino)tetrahydro-2H-pyran-4-yl)acrylamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3S,4S)-3-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl]amino}oxan-4-yl]propanamide, SULFATE ION | | Authors: | Lin, X, Smaill, J.B, Squire, C.J, Yosaatmadja, Y. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rotational Freedom, Steric Hindrance, and Protein Dynamics Explain BLU554 Selectivity for the Hinge Cysteine of FGFR4.
Acs Med.Chem.Lett., 10, 2019
|
|
8H9I
 
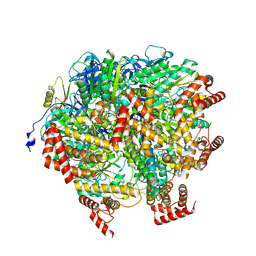 | | Human ATP synthase F1 domain, state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit O, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9Q
 
 | | Human ATP synthase state 3b subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9R
 
 | | Human ATP synthase state 3b subregion 2 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase subunit d, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
