6MBJ
 
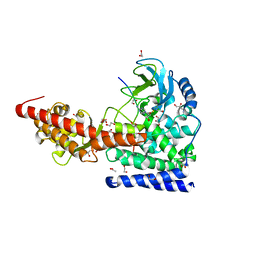 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, P21 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MG1
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta, ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MBK
 
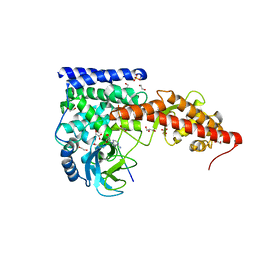 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, First P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MBL
 
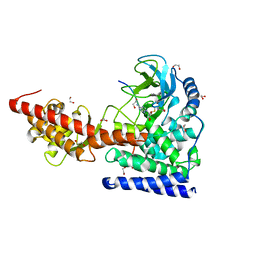 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, Second P212121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
6MG3
 
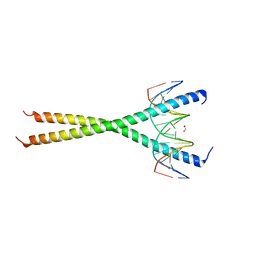 | | V285A Mutant of the C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
6MG2
 
 | | C-terminal bZIP domain of human C/EBPbeta with 16bp Methylated Oligonucleotide Containing Consensus Recognition Sequence-C2221 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 16-bp methylated oligonucleotide, CCAAT/enhancer-binding protein beta | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.928 Å) | | Cite: | Structural basis for effects of CpA modifications on C/EBP beta binding of DNA.
Nucleic Acids Res., 47, 2019
|
|
7XW7
 
 | | TSHR-K1-70 complex | | Descriptor: | K1-70 scFv, Thyrotropin receptor | | Authors: | Duan, J, Xu, P, Luan, X, Ji, Y, Yuan, Q, He, X, Ye, J, Cheng, X, Jiang, H, Zhang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-05-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Hormone- and antibody-mediated activation of the thyrotropin receptor.
Nature, 609, 2022
|
|
7XW6
 
 | | TSHR-Gs-M22 antibody-ML109 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Duan, J, Xu, P, Luan, X, Ji, Y, Yuan, Q, He, X, Ye, J, Cheng, X, Jiang, H, Zhang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-05-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Hormone- and antibody-mediated activation of the thyrotropin receptor.
Nature, 609, 2022
|
|
8FS1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8FS2
 
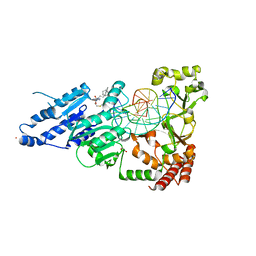 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11b (YD907) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{3-[N'-(cyclohexylmethyl)carbamimidamido]propyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
4RNW
 
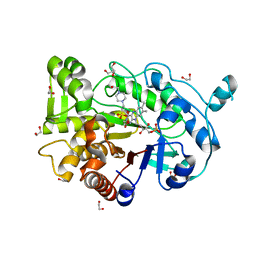 | | Truncated version of the G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4QW2
 
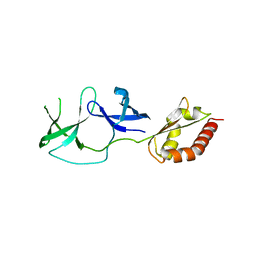 | | FMRP N-terminal domain (R138Q) | | Descriptor: | 1,2-ETHANEDIOL, Fragile X mental retardation protein 1, LEAD (II) ION | | Authors: | Myrick, L.K, Hashimoto, H, Cheng, X, Warren, S.T. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.
Hum.Mol.Genet., 24, 2015
|
|
5T00
 
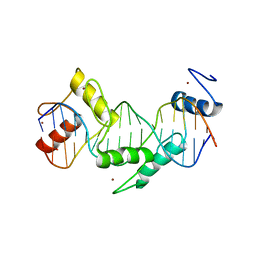 | | Human CTCF ZnF3-7 and methylated DNA complex | | Descriptor: | DNA (5'-GCCAGCAGGGGG(5CM)GCTA-3'), DNA (5'-TAG(5CM)GCCCCCTGCTGGC-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
4RNX
 
 | | K154 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1 | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
5T01
 
 | | Human c-Jun DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*GP*AP*(5CM)P*GP*AP*GP*TP*CP*AP*TP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*CP*TP*AP*TP*GP*AP*CP*TP*CP*GP*TP*CP*CP*AP*T)-3'), Transcription factor AP-1 | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
4QVZ
 
 | | FMRP N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Fragile X mental retardation protein 1 | | Authors: | Myrick, L.K, Hashimoto, H, Cheng, X, Warren, S.T. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.
Hum.Mol.Genet., 24, 2015
|
|
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
4RNV
 
 | | G303 Circular Permutation of Old Yellow Enzyme with the Inhibitor p-Hydroxybenzaldehyde | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, P-HYDROXYBENZALDEHYDE | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RNU
 
 | | G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
5T0U
 
 | | CTCF ZnF2-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
4F7Z
 
 | | Conformational dynamics of exchange protein directly activated by cAMP | | Descriptor: | GLYCEROL, Rap guanine nucleotide exchange factor 4 | | Authors: | White, M.A, Tsalkova, T.N, Mei, F.C, Liu, T, Woods, V.L, Cheng, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analyses of a constitutively active mutant of exchange protein directly activated by cAMP.
Plos One, 7, 2012
|
|
7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
2PVI
 
 | | PVUII ENDONUCLEASE COMPLEXED TO AN IODINATED COGNATE DNA | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*CP*AP*GP*(C38)P*TP*GP*GP*TP*C)-3'), TYPE II RESTRICTION ENZYME PVUII | | Authors: | Horton, J, Cheng, X. | | Deposit date: | 1998-11-01 | | Release date: | 1999-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | How is modification of the DNA substrate recognized by the PvuII restriction endonuclease?
J.Biol.Chem., 379, 1998
|
|
7N5S
 
 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 6) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7N5T
 
 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 5) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
