3I18
 
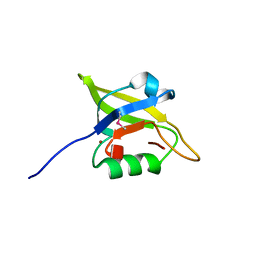 | | Crystal Structure of the PDZ domain of the SdrC-like protein (Lmo2051) from Listeria monocytogenes, Northeast Structural Genomics Consortium Target LmR166B | | Descriptor: | BROMIDE ION, Lmo2051 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Janjua, J, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-25 | | Release date: | 2009-07-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target LmR166B
To be Published
|
|
5L03
 
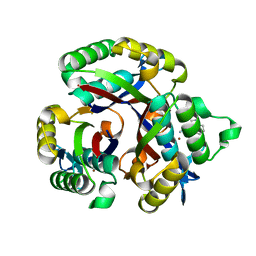 | | Crystal structure of 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE Synthase from BURKHOLDERIA PSEUDOMALLEI bound to L-tryptophan hydroxamate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, N-hydroxy-L-tryptophanamide, ZINC ION | | Authors: | Blain, J.M, Ghose, D, Gorman, J.L, Goshu, G.M, Ranieri, G, Zhao, L, Bode, B, Meganathan, R, Walter, R.L, Hagen, T.J, Horn, J.R. | | Deposit date: | 2016-07-26 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Synthesis and Characterization of the Burkholderia pseudomallei IspF Inhibitor L-tryptophan hydroxamate
To Be Published
|
|
3IHK
 
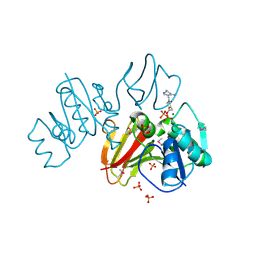 | | Crystal Structure of thiamin pyrophosphokinase from S.mutans, Northeast Structural Genomics Consortium Target SmR83 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Vorobiev, S, Mao, M, Xiao, R, Ciccosanti, C, Maglaqui, M, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Northeast Structural Genomics Consortium Target SmR83
To be published
|
|
3HXJ
 
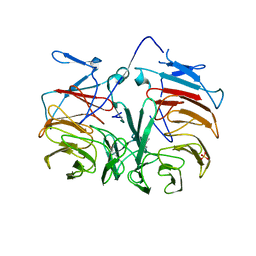 | | Crystal Structure of Pyrrolo-quinoline quinone (PQQ_DH) from Methanococcus maripaludis, Northeast Structural Genomics Consortium Target MrR86 | | Descriptor: | Pyrrolo-quinoline quinone, SULFATE ION | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target MrR86
To be Published
|
|
3JVN
 
 | | Crystal Structure of the acetyltransferase VF_1542 from Vibrio fischeri, Northeast Structural Genomics Consortium Target VfR136 | | Descriptor: | Acetyltransferase, SULFATE ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-17 | | Release date: | 2009-09-29 | | Last modified: | 2019-08-07 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Northeast Structural Genomics Consortium Target VfR136
To be Published
|
|
3HXK
 
 | | Crystal Structure of a sugar hydrolase (YeeB) from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR108 | | Descriptor: | Sugar hydrolase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-20 | | Release date: | 2009-07-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of a sugar hydrolase (YeeB) from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR108
To be Published
|
|
5H1B
 
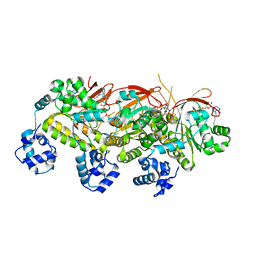 | | Human RAD51 presynaptic complex | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, MAGNESIUM ION, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
5H1C
 
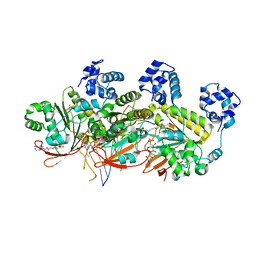 | | Human RAD51 post-synaptic complexes | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | Deposit date: | 2016-10-08 | | Release date: | 2016-12-21 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
2I4R
 
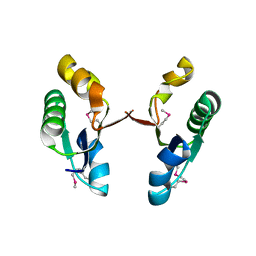 | | Crystal structure of the V-type ATP synthase subunit F from Archaeoglobus fulgidus. NESG target GR52A. | | Descriptor: | V-type ATP synthase subunit F | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Zhao, L, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-22 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the V-type ATP synthase subunit F from Archaeoglobus fulgidus
To be Published
|
|
3CPK
 
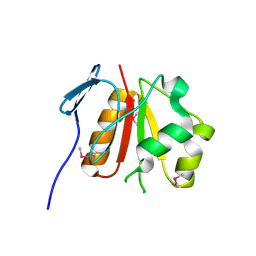 | | Crystal structure of the Q7W7N7_BORPA protein from Bordetella parapertussis. Northeast Structural Genomics Consortium target BeR31 | | Descriptor: | Uncharacterized protein Q7W7N7_BORPA | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Zhao, L, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Q7W7N7_BORPA protein from Bordetella parapertussis.
To be Published
|
|
3CQ9
 
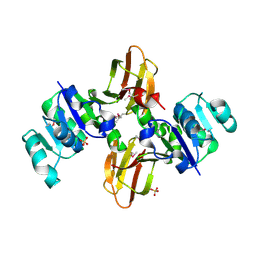 | | Crystal structure of the lp_1622 protein from Lactobacillus plantarum. Northeast Structural Genomics Consortium target LpR114 | | Descriptor: | SULFATE ION, Uncharacterized protein lp_1622 | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Zhao, L, Maglaqui, M, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-04-02 | | Release date: | 2008-04-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the lp_1622 protein from Lactobacillus plantarum.
To be Published
|
|
2KKZ
 
 | | Solution NMR structure of the monomeric W187R mutant of A/Udorn NS1 effector domain. Northeast Structural Genomics target OR8C[W187R]. | | Descriptor: | Non-structural protein NS1 | | Authors: | Aramini, J.M, Ma, L, Lee, H, Zhao, L, Cunningham, K, Ciccosanti, C, Janjua, H, Fang, Y, Xiao, R, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the monomeric W187R mutant of A/Udorn NS1 effector domain. Northeast Structural Genomics target OR8C[W187R].
To be Published
|
|
4DLH
 
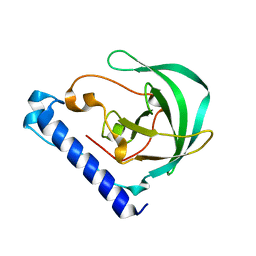 | | Crystal Structure of the protein Q9HRE7 from Halobacterium salinarium at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HsR50 | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
2KFP
 
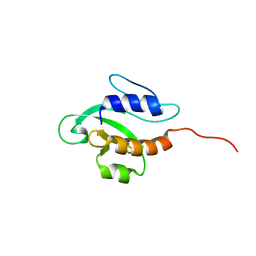 | | Solution NMR structure of PSPTO_3016 from Pseudomonas syringae. Northeast Structural Genomics Consortium target PsR293. | | Descriptor: | PSPTO_3016 protein | | Authors: | Feldmann, E.A, Ramelot, T.A, Zhao, L, Hamilton, K, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and X-ray crystal structures of Pseudomonas syringae Pspto_3016 from protein domain family PF04237 (DUF419) adopt a "double wing" DNA binding motif.
J.Struct.Funct.Genom., 13, 2012
|
|
3MEL
 
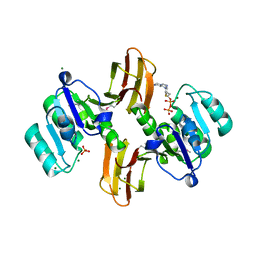 | | Crystal Structure of Thiamin pyrophosphokinase family protein from Enterococcus faecalis, Northeast Structural Genomics Consortium Target EfR150 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Kuzin, A, Abasidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Northeast Structural Genomics Consortium Target EfR150
To be Published
|
|
3MGD
 
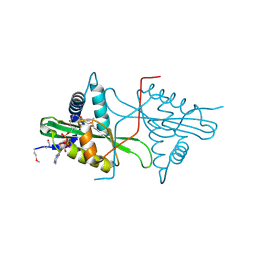 | | Crystal Structure of predicted acetyltransferase with acetyl-CoA from Clostridium acetobutylicum at the resolution 1.9A, Northeast Structural Genomics Consortium Target CaR165 | | Descriptor: | ACETYL COENZYME *A, Predicted acetyltransferase | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-28 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target CaR165
To be published
|
|
8DPX
 
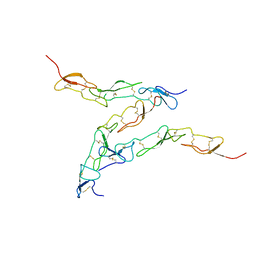 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
1T6M
 
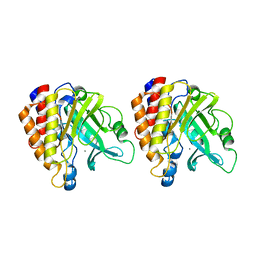 | | X-ray Structure of the R70D PI-PLC enzyme: Insight into the role of calcium and surrounding amino acids on active site geometry and catalysis. | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, CALCIUM ION | | Authors: | Apiyo, D, Zhao, L, Tsai, M.-D, Selby, T.L. | | Deposit date: | 2004-05-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | X-ray Structure of the R69D Phosphatidylinositol-Specific Phospholipase C Enzyme: Insight into the Role of Calcium and Surrounding Amino Acids in Active Site Geometry and Catalysis.
Biochemistry, 44, 2005
|
|
2ICP
 
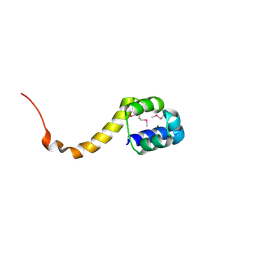 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 4.0. Northeast Structural Genomics Consortium TARGET ER390. | | Descriptor: | MAGNESIUM ION, antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the bacterial antitoxin HigA from Escherichia coli.
To be Published
|
|
4Z5V
 
 | |
2Q0Z
 
 | | Crystal structure of Q9P172/Sec63 from Homo sapiens. Northeast Structural Genomics Target HR1979. | | Descriptor: | Protein PRO2281 | | Authors: | Benach, J, Neely, H, Abashidze, M, Edstrom, W.C, Seetharaman, J, Zhao, L, Fang, Y, Cunningham, K, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-05-23 | | Release date: | 2007-06-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Q9P172/Sec63 from Homo sapiens.
To be Published
|
|
3I1H
 
 | | Crystal structure of human BFL-1 in complex with BAK BH3 peptide | | Descriptor: | Apoptosis regulator BAK, Protein BFL-1 | | Authors: | Guan, R, Xiao, R, Zhao, L, Acton, T, White, E, Gelinas, C, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | crystal structure of human BFL-1 in complex with BAK BH3 peptide
To be Published
|
|
2NW7
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferric heme. Northeast Structural Genomics Target XcR13 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Bruckmann, C, Thackray, S.J, Seetharaman, J, Tucker, T, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Chapman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2O8S
 
 | | X-ray Crystal Structure of Protein AGR_C_984 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR120. | | Descriptor: | AGR_C_984p, SULFATE ION | | Authors: | Seetharaman, J, Forouhar, F, Su, M, Zhao, L, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Hypothetical Protein AGR_C_984 from Agrobacterium tumefaciens, Northeast Structural Genomics Target AtR120.
To be Published
|
|
2NW8
 
 | | Crystal Structure of Tryptophan 2,3-dioxygenase (TDO) from Xanthomonas campestris in complex with ferrous heme and tryptophan. Northeast Structural Genomics Target XcR13. | | Descriptor: | MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, ... | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Bruckmann, C, Thackray, S.J, Seetharaman, J, Ho, C.K, Ma, L.C, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
