2OO5
 
 | | Structure of transhydrogenase (dI.H2NADH)2(dIII.NADP+)1 asymmetric complex | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Bhakta, T, Jackson, J.B. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Biochemistry, 46, 2007
|
|
2OOR
 
 | | Structure of transhydrogenase (dI.NAD+)2(dIII.H2NADPH)1 asymmetric complex | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Bhakta, T, Jackson, J.B. | | Deposit date: | 2007-01-26 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Biochemistry, 46, 2007
|
|
1E3T
 
 | | Solution Structure of the NADP(H) binding Component (dIII) of Proton-Translocating Transhydrogenase from Rhodospirillum rubrum | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE (SUBUNIT BETA) | | Authors: | Jeeves, M, Smith, K.J, Quirk, P.G, Cotton, N.P.J, Jackson, J.B. | | Deposit date: | 2000-06-22 | | Release date: | 2000-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Nadp(H)-Binding Component (Diii) of Proton-Translocating Transhydrogenase from Rhodospirillum Rubrum
Biochim.Biophys.Acta, 1459, 2000
|
|
1HZZ
 
 | | THE ASYMMETRIC COMPLEX OF THE TWO NUCLEOTIDE-BINDING COMPONENTS (DI, DIII) OF PROTON-TRANSLOCATING TRANSHYDROGENASE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTON-TRANSLOCATING NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE SUBUNIT PNTAA, ... | | Authors: | Cotton, N.P.J, White, S.A, Peake, S.J, McSweeney, S, Jackson, J.B. | | Deposit date: | 2001-01-27 | | Release date: | 2001-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of an asymmetric complex of the two nucleotide binding components of proton-translocating transhydrogenase.
Structure, 9, 2001
|
|
6YCL
 
 | | Crystal structure of GcoA T296G bound to p-vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCK
 
 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCO
 
 | | Crystal structure of GcoA F169S bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCP
 
 | | Crystal structure of GcoA F169V bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCT
 
 | | Crystal structure of GcoA F169A_T296S bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Cytochrome P450, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCN
 
 | | Crystal structure of GcoA F169A bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCI
 
 | | Crystal structure of GcoA T296G bound to guaiacol | | Descriptor: | Aromatic O-demethylase, cytochrome P450 subunit, Guaiacol, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
7R9X
 
 | |
6JJL
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-ARG-LYS-LEU, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy.
Commun Biol, 3, 2020
|
|
6JJK
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | CYS-TYR-TYR-LYS-ILE, Periplasmic serine endoprotease DegP | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
6JJO
 
 | | Crystal structure of the DegP dodecamer with a modulator | | Descriptor: | Periplasmic serine endoprotease DegP, TMB-CYRKL modulator | | Authors: | Cho, H, Choi, Y, Lee, H.H, Kim, S. | | Deposit date: | 2019-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.157 Å) | | Cite: | Over-activation of a nonessential bacterial protease DegP as an antibiotic strategy
Commun Biol, 3, 2020
|
|
8E72
 
 | | Treponema lecithinolyticum beta-glucuronidase in complex with a ciprofloxacin-glucuronide conjugate | | Descriptor: | 3-carboxy-1-cyclopropyl-6-fluoro-7-(4-beta-D-glucopyranuronosyl-3,4-dihydropyrazin-1(2H)-yl)-4-oxo-1,4-dihydroquinoline, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-08-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
6PBO
 
 | | Staphylococcus aureus Dihydrofolate reductase in complex with NADPH and UCP1232 | | Descriptor: | (4-{6-[(2S)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-2H-1,3-benzodioxol-4-yl}phenyl)acetic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Reeve, S.M, Wright, D.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Toward Broad Spectrum Dihydrofolate Reductase Inhibitors Targeting Trimethoprim Resistant Enzymes Identified in Clinical Isolates of Methicillin ResistantStaphylococcus aureus.
Acs Infect Dis., 5, 2019
|
|
6P9Z
 
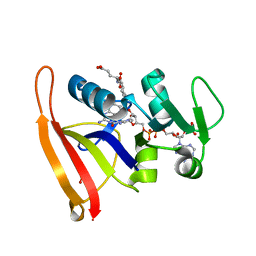 | |
8DHL
 
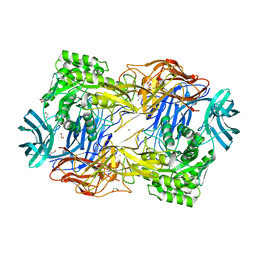 | | Tannerella forsythia beta-glucuronidase (L2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHV
 
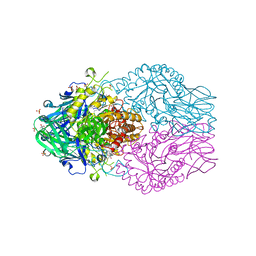 | | Treponema lecithinolyticum beta-glucuronidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHE
 
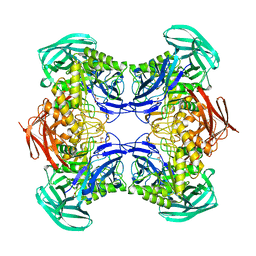 | |
8DHW
 
 | | Treponema lecithinolyticum beta-glucuronidase in complex with a UNC4917-glucuronide conjugate | | Descriptor: | 4-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-2,7-bis(methylamino)pyrido[3',2':4,5]thieno[3,2-d]pyrimidine, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8SBG
 
 | |
8SIJ
 
 | | Crystal structure of F. varium tryptophanase | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Tryptophanase 1, ... | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
8SL7
 
 | | Butyricicoccus sp. BIOML-A1 tryptophanase complex with (3S) ALG-05 | | Descriptor: | (E)-3-[(3S)-3-chloro-2-oxo-2,3-dihydro-1H-indol-3-yl]-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, Tryptophanase | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
