2KA9
 
 | | Solution structure of PSD-95 PDZ12 complexed with cypin peptide | | Descriptor: | Disks large homolog 4, cypin peptide | | Authors: | Wang, W.N, Weng, J.W, Zhang, X, Liu, M.L, Zhang, M.J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Creating conformational entropy by increasing interdomain mobility in ligand binding regulation: a revisit to N-terminal tandem PDZ domains of PSD-95
J.Am.Chem.Soc., 131, 2009
|
|
1BR0
 
 | | THREE DIMENSIONAL STRUCTURE OF THE N-TERMINAL DOMAIN OF SYNTAXIN 1A | | Descriptor: | PROTEIN (SYNTAXIN 1-A) | | Authors: | Fernandez, I, Ubach, J, Dubulova, I, Zhang, X, Sudhof, T.C, Rizo, J. | | Deposit date: | 1998-08-25 | | Release date: | 1998-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A.
Cell(Cambridge,Mass.), 94, 1998
|
|
2AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 20 STRUCTURES | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-28 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
3MUS
 
 | | 2A Resolution Structure of Rat Type B Cytochrome b5 | | Descriptor: | Cytochrome b5 type B, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X, Benson, D.R. | | Deposit date: | 2010-05-03 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accommodating a Non-Conservative Internal Mutation by Water-Mediated Hydrogen-Bonding Between beta-Sheet Strands: A Comparison of Human and Rat Type B (Mitochondrial) Cytochrome b5
Biochemistry, 50, 2011
|
|
1D59
 
 | | CRYSTAL STRUCTURE OF 4-STRANDED OXYTRICHA TELOMERIC DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3') | | Authors: | Kang, C, Zhang, X, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1992-02-25 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of four-stranded Oxytricha telomeric DNA.
Nature, 356, 1992
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2013-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
2J48
 
 | |
5EZK
 
 | | RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex. | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darbari, V.C, Yang, Y, Lu, D, Zhang, N, Glyde, R, Wang, Y, Murakami, K.S, Buck, M, Zhang, X. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | TRANSCRIPTION. Structures of the RNA polymerase- Sigma 54 reveal new and conserved regulatory strategies.
Science, 349, 2015
|
|
5YCO
 
 | | Complex structure of PCNA with UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
2IPA
 
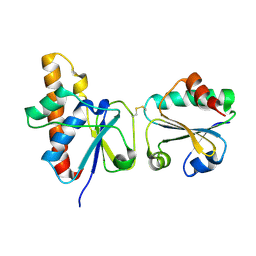 | | solution structure of Trx-ArsC complex | | Descriptor: | Protein arsC, Thioredoxin | | Authors: | Jin, C, Hu, Y, Li, Y, Zhang, X. | | Deposit date: | 2006-10-12 | | Release date: | 2007-02-13 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis.
J.Biol.Chem., 282, 2007
|
|
3F8I
 
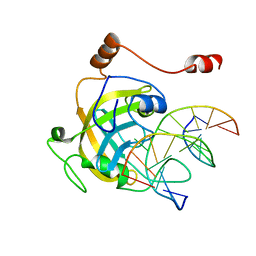 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group P21 | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3FDE
 
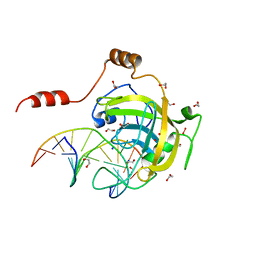 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3F8J
 
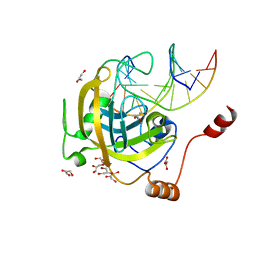 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
1U2Z
 
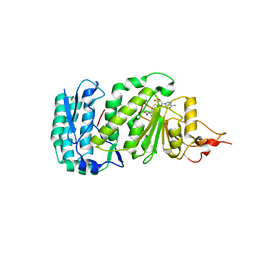 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
6BP7
 
 | | Recombinant major vault protein [Rattus norvegicus] structure in solution: conformation 2 | | Descriptor: | Major vault protein | | Authors: | Ding, K, Zhang, X, Mrazek, J, Kickhoefer, V.A, Lai, M, Ng, H.L, Yang, O.O, Rome, L.H, Zhou, Z.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Solution Structures of Engineered Vault Particles.
Structure, 26, 2018
|
|
3KV9
 
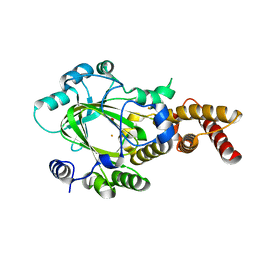 | | Structure of KIAA1718 Jumonji domain | | Descriptor: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6BZE
 
 | | Cryo-EM structure of BCL10 CARD filament | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | David, L, Li, Y, Ma, J, Garner, E, Zhang, X, Wu, H. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4AM6
 
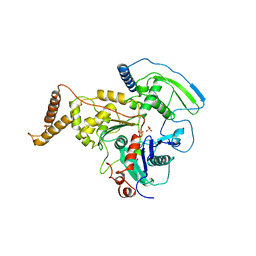 | | C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, SULFATE ION | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AM7
 
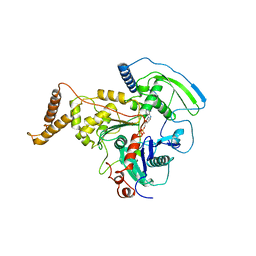 | | ADP-BOUND C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6BP8
 
 | | Recombinant major vault protein [Rattus norvegicus] structure in solution: conformation 1 | | Descriptor: | Major vault protein | | Authors: | Ding, K, Zhang, X, Mrazek, J, Kickhoefer, V.A, Lai, M, Ng, H.L, Yang, O.O, Rome, L.H, Zhou, Z.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Solution Structures of Engineered Vault Particles.
Structure, 26, 2018
|
|
8II0
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(3-(3-chlorophenyl)isoxazol-5-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[3-(3-chlorophenyl)-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8IHZ
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(1-(3-(4-chlorophenyl)propyl)-1H-1,2,3-triazol-4-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[1-[3-(4-chlorophenyl)propyl]-1,2,3-triazol-4-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 2024
|
|
3J6C
 
 | | Cryo-EM structure of MAVS CARD filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | Deposit date: | 2014-02-04 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2GZY
 
 | |
