8V3S
 
 | | Structure of CCP5 class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4K
 
 | | CCP5 in complex with microtubules class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3P
 
 | | CCP5 in complex with Glu-P-peptide 2 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, Tubulin beta-2A chain, ZINC ION | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3O
 
 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4M
 
 | | CCP5 in complex with microtubules class3 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3Q
 
 | | Structure of CCP5 class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4L
 
 | | CCP5 in complex with microtubules class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3M
 
 | | CCP5 apo structure | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3R
 
 | | Structure of CCP5 class2 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8XEG
 
 | |
8YU7
 
 | | Cryo-EM structure of CXCR4 tetramer | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL | | Authors: | Liu, A, Liu, Y. | | Deposit date: | 2024-03-26 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis of CXCR4 assembly and regulation.
Cell Rep, 44, 2025
|
|
8YYX
 
 | | Cryo-EM structure of OXGR1 bound to leukotriene E4 and Gq proteins | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, 2-oxoglutarate receptor 1, CHOLESTEROL, ... | | Authors: | Liu, A, Liu, Y. | | Deposit date: | 2024-04-04 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of OXGR1 bound to leukotriene E4 and Gq proteins
To Be Published
|
|
8YYW
 
 | |
7XC4
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | Descriptor: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | Authors: | Li, J, Liu, Y, Gao, J, Ruan, K. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
8J4I
 
 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
7DPD
 
 | | Human MCM9 N-terminal domain | | Descriptor: | DNA helicase MCM9, SODIUM ION, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
7DP3
 
 | | Human MCM8 N-terminal domain | | Descriptor: | DNA helicase MCM8, ZINC ION | | Authors: | Li, J, Liu, L, Liu, Y. | | Deposit date: | 2020-12-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural study of the N-terminal domain of human MCM8/9 complex.
Structure, 29, 2021
|
|
6JWE
 
 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
7YEH
 
 | | Cryo-EM structure of human OGT-OGA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, ... | | Authors: | Lu, P, Liu, Y, Yu, H, Gao, H. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
7YEA
 
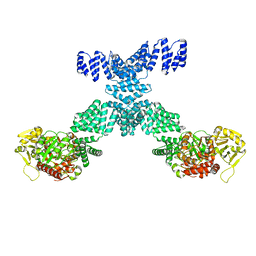 | | Human O-GlcNAc transferase Dimer | | Descriptor: | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gao, H, Lu, P, Liu, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
7X8M
 
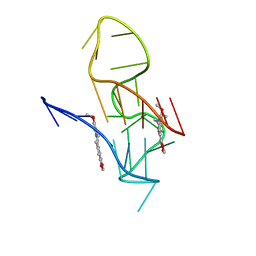 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | Descriptor: | BERBERINE, DNA (24-MER) | | Authors: | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
6B0V
 
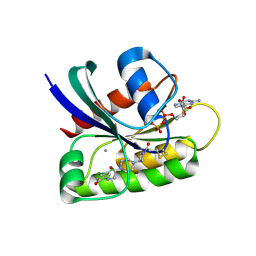 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8ZDY
 
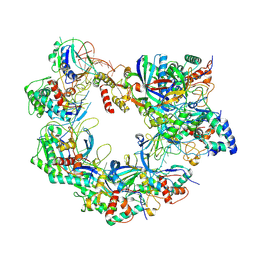 | | Cryo-EM structure of Cas8-HNH system at target free state | | Descriptor: | RNA (58-MER), a protein | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8ZNR
 
 | | Cryo-EM structure of Cas8-HNH system at ssDNA-bound state | | Descriptor: | DNA (35-MER), RNA (56-MER), protein structure | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
