8DMD
 
 | |
6VKM
 
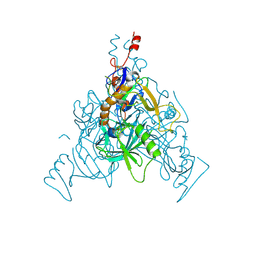 | | Crystal Structure of Stabilized GP from Makona Variant of Ebola Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Virion spike glycoprotein | | Authors: | Gilman, M.S.A, Rutten, L, Langedijk, J.P.M, McLellan, J.S. | | Deposit date: | 2020-01-21 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Based Design of Prefusion-Stabilized Filovirus Glycoprotein Trimers.
Cell Rep, 30, 2020
|
|
2P3T
 
 | |
6MLM
 
 | | H7 HA0 in complex with Fv from H7.5 IgG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain Fv of H7.5 Fab, Hemagglutinin HA1 chain, ... | | Authors: | Pallesen, J, Turner, H.L, Ward, A.B. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains.
PLoS Biol., 17, 2019
|
|
7S4B
 
 | |
7S3S
 
 | |
7S3K
 
 | |
5FHA
 
 | |
5FHB
 
 | |
5T45
 
 | | Chicken smooth muscle myosin motor domain co-crystallized with the specific CK-571 inhibitor, MgADP.BeFx form | | Descriptor: | 4-{[(2-chloro-3-fluorobenzyl)carbamoyl](methyl)amino}-3,4-dideoxy-5-O-(isoquinolin-3-ylcarbamoyl)-D-erythro-pentitol, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Sirigu, S, Planelles-Herrero, V.J, Hartman, J, Houdusse, A. | | Deposit date: | 2016-08-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Highly selective inhibition of myosin motors provides the basis of potential therapeutic application.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7A4N
 
 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(S-closed trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-04 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
5FCS
 
 | | Diabody | | Descriptor: | Diabody, SULFATE ION | | Authors: | Mosyak, L, Root, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-14 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Development of PF-06671008, a Highly Potent Anti-P-cadherin/Anti-CD3 Bispecific DART Molecule with Extended Half-Life for the Treatment of Cancer.
Antibodies, 5, 2016
|
|
6I9R
 
 | | Large subunit of the human mitochondrial ribosome in complex with Virginiamycin M and Quinupristin | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Modelska, A, Aibara, S, Amunts, A. | | Deposit date: | 2018-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth.
Cell Rep, 35, 2021
|
|
7AD1
 
 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(One up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein,Spike glycoprotein,Envelope glycoprotein,SARS-CoV-2 S protein | | Authors: | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | Deposit date: | 2020-09-14 | | Release date: | 2020-11-04 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
4MSL
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF40431 | | Descriptor: | N-[(7-hydroxy-4-methyl-2-oxo-2H-chromen-8-yl)methyl]-L-leucine, Sortilin, TETRAETHYLENE GLYCOL, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the first small-molecule ligand of the neuronal receptor sortilin and structure determination of the receptor-ligand complex.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5FHC
 
 | |
4N7E
 
 | | Crystal structure of the Vps10p domain of human sortilin/NTS3 in complex with AF38469 | | Descriptor: | 2-[(6-methylpyridin-2-yl)carbamoyl]-5-(trifluoromethyl)benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, ... | | Authors: | Andersen, J.L, Strandbygaard, D, Thirup, S. | | Deposit date: | 2013-10-15 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The identification of AF38469: An orally bioavailable inhibitor of the VPS10P family sorting receptor Sortilin.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2P3U
 
 | |
1VGE
 
 | |
7TJE
 
 | | Bacteriophage Q beta capsid protein A38K | | Descriptor: | Minor capsid protein A1 | | Authors: | Jin, X. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Alternative Assembly of Q beta Virus-like Particles
To Be Published
|
|
7TJG
 
 | |
7TJM
 
 | |
7TJD
 
 | |
5OMZ
 
 | |
6MBP
 
 | | GLP Methyltransferase with Inhibitor EML741- P3121 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-7-methoxy-N-[1-(propan-2-yl)piperidin-4-yl]-8-[3-(pyrrolidin-1-yl)propoxy]-3H-1,4-benzodiazepin-5-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Discovery of a Novel Chemotype of Histone Lysine Methyltransferase EHMT1/2 (GLP/G9a) Inhibitors: Rational Design, Synthesis, Biological Evaluation, and Co-crystal Structure.
J. Med. Chem., 62, 2019
|
|
