8YBR
 
 | | Choline transporter BetT | | Descriptor: | BCCT family transporter | | Authors: | Yang, T.J, Nian, Y.W, Lin, H.J, Li, J, Zhang, J.R, Fan, M.R. | | Deposit date: | 2024-02-16 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structure and mechanism of the osmoregulated choline transporter BetT.
Sci Adv, 10, 2024
|
|
5SV7
 
 | | The Crystal structure of a chaperone | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wang, P, Li, J, Sha, B. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8XI3
 
 | | Structure of mouse SCMC-14-3-3gama complex | | Descriptor: | 14-3-3 protein gamma, NACHT, LRR and PYD domains-containing protein 5, ... | | Authors: | Chi, P, Han, Z, Jiao, H, Li, J, Wang, X, Hu, H, Deng, D. | | Deposit date: | 2023-12-19 | | Release date: | 2024-10-02 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The subcortical maternal complex modulates the cell cycle during early mammalian embryogenesis via 14-3-3.
Nat Commun, 15, 2024
|
|
7SGL
 
 | | DNA-PK complex of DNA end processing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, Hairpin_1, ... | | Authors: | Liu, L, Li, J, Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2021-10-06 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
2R5C
 
 | | Aedes Kynurenine Aminotransferase in Complex with Cysteine | | Descriptor: | Kynurenine aminotransferase, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Li, J. | | Deposit date: | 2007-09-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
Biochemistry, 47, 2008
|
|
6J73
 
 | | Crystal structure of IniA from Mycobacterium smegmatis | | Descriptor: | Isoniazid inducible gene protein IniA | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
5JHH
 
 | | Crystal structure of the ternary complex between the human RhoA, its inhibitor and the DH/PH domain of human ARHGEF11 | | Descriptor: | 3-{3-[ethyl(quinolin-2-yl)amino]phenyl}propanoic acid, GLYCEROL, Rho guanine nucleotide exchange factor 11, ... | | Authors: | Lv, Z, Wang, R, Ma, L, Miao, Q, Wu, J, Yan, Z, Li, J, Miao, L, Wang, F. | | Deposit date: | 2016-04-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of a small GTPase RhoA bound with its inhibitor and PDZRhoGEF
To Be Published
|
|
2QTW
 
 | | The Crystal Structure of PCSK9 at 1.9 Angstroms Resolution Reveals structural homology to Resistin within the C-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Hampton, E.N, Knuth, M.W, Li, J, Harris, J.L, Lesley, S.A, Spraggon, G. | | Deposit date: | 2007-08-02 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R2N
 
 | | The crystal structure of human kynurenine aminotransferase II in complex with kynurenine | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, ... | | Authors: | Han, Q, Robinson, H, Li, J. | | Deposit date: | 2007-08-27 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human kynurenine aminotransferase II.
J.Biol.Chem., 283, 2008
|
|
2R5E
 
 | | Aedes kynurenine aminotransferase in complex with glutamine | | Descriptor: | Kynurenine aminotransferase, N~2~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-GLUTAMINE | | Authors: | Han, Q, Gao, Y.G, Robinson, H, Li, J. | | Deposit date: | 2007-09-03 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the mechanism of substrate specificity of aedes kynurenine aminotransferase.
Biochemistry, 47, 2008
|
|
5U2U
 
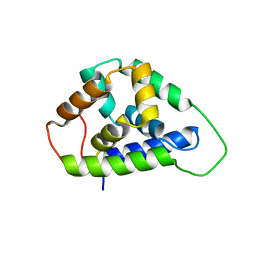 | |
2Q8F
 
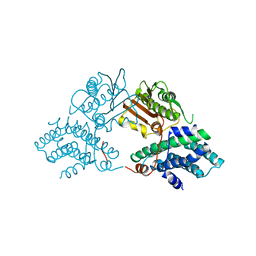 | | Structure of pyruvate dehydrogenase kinase isoform 1 | | Descriptor: | POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
3RHZ
 
 | |
2QLR
 
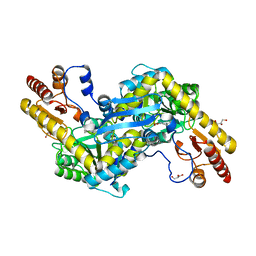 | |
5U2L
 
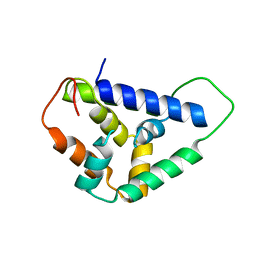 | | Crystal structure of the Hsp104 N-terminal domain from Candida albicans | | Descriptor: | Heat shock protein 104 | | Authors: | Wang, P, Li, J, Sha, B. | | Deposit date: | 2016-11-30 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6555 Å) | | Cite: | Crystal structures of Hsp104 N-terminal domains from Saccharomyces cerevisiae and Candida albicans suggest the mechanism for the function of Hsp104 in dissolving prions.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1OLS
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1J48
 
 | | Crystal Structure of Apo-C1027 | | Descriptor: | Apoprotein of C1027 | | Authors: | Chen, Y, Li, J, Liu, Y, Bartlam, M, Gao, Y, Jin, L, Tang, H, Shao, Y, Zhen, Y, Rao, Z. | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo-C1027 and Computer Modeling Analysis of C1027 Chromophore- Protein Complex
To be published
|
|
2Q8G
 
 | | Structure of pyruvate dehydrogenase kinase isoform 1 in complex with glucose-lowering drug AZD7545 | | Descriptor: | 4-[(3-CHLORO-4-{[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]AMINO}PHENYL)SULFONYL]-N,N-DIMETHYLBENZAMIDE, POTASSIUM ION, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1 | | Authors: | Kato, M, Li, J, Chuang, J.L, Chuang, D.T. | | Deposit date: | 2007-06-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol.
Structure, 15, 2007
|
|
8J1I
 
 | | Crystal Structure of EphA8/SASH1 Complex | | Descriptor: | Ephrin type-A receptor 8, SAM and SH3 domain-containing protein 1 | | Authors: | Liu, W, Li, J, Ding, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | SASH1: A Novel Eph Receptor Partner and Insights into SAM-SAM Interactions.
J.Mol.Biol., 435, 2023
|
|
7VLN
 
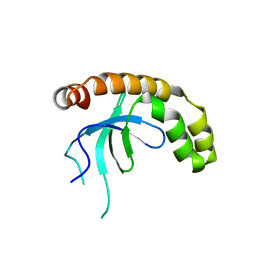 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | Descriptor: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | Authors: | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7W8N
 
 | | Microbial Hormone-sensitive lipase E53 wild type | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, ... | | Authors: | Yang, X, Li, Z, Xu, X, Li, J. | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-23 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism and Structural Insights Into a Novel Esterase, E53, Isolated From Erythrobacter longus .
Front Microbiol, 12, 2021
|
|
6IY1
 
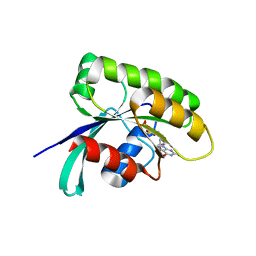 | | Structure of human Ras-related protein Rab11 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-11A | | Authors: | Ma, P, Li, S, Li, J. | | Deposit date: | 2018-12-12 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of human Ras-related protein Rab11
To Be Published
|
|
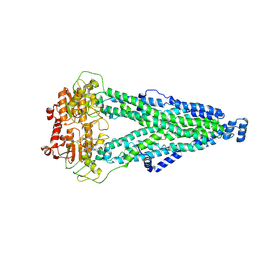
8XSS
 
 | |
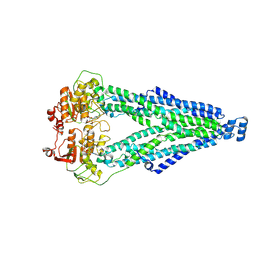
8XSR
 
 | |
